[English] 日本語
 Yorodumi
Yorodumi- PDB-7x6r: Crystal structure of actinomycin D-echinomycin-d(AGCACGT/ACGGGCT)... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7x6r | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of actinomycin D-echinomycin-d(AGCACGT/ACGGGCT) complex | ||||||||||||
 Components Components |
| ||||||||||||
 Keywords Keywords | DNA/ANTIBIOTIC / A:G mismatch / Single-strand DNA distortion / Actinomycin D / Echinomycin / mismatch binding drug / DNA / DNA-ANTIBIOTIC complex | ||||||||||||
| Function / homology | Actinomycin D / Echinomycin / : / 2-CARBOXYQUINOXALINE / DNA Function and homology information Function and homology information | ||||||||||||
| Biological species |  Streptomyces (bacteria) Streptomyces (bacteria)synthetic construct (others) | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.89 Å MOLECULAR REPLACEMENT / Resolution: 1.89 Å | ||||||||||||
 Authors Authors | Kao, S.H. / Satange, R.B. / Hou, M.H. | ||||||||||||
| Funding support |  Taiwan, 2items Taiwan, 2items
| ||||||||||||
 Citation Citation |  Journal: Nucleic Acids Res. / Year: 2022 Journal: Nucleic Acids Res. / Year: 2022Title: Staggered intercalation of DNA duplexes with base-pair modulation by two distinct drug molecules induces asymmetric backbone twisting and structure polymorphism. Authors: Satange, R. / Kao, S.H. / Chien, C.M. / Chou, S.H. / Lin, C.C. / Neidle, S. / Hou, M.H. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7x6r.cif.gz 7x6r.cif.gz | 41.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7x6r.ent.gz pdb7x6r.ent.gz | 27.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7x6r.json.gz 7x6r.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/x6/7x6r https://data.pdbj.org/pub/pdb/validation_reports/x6/7x6r ftp://data.pdbj.org/pub/pdb/validation_reports/x6/7x6r ftp://data.pdbj.org/pub/pdb/validation_reports/x6/7x6r | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7x97C  7x9fC  7xdjC  7dq0S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 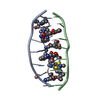
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||
| Components on special symmetry positions |
|
- Components
Components
-DNA chain , 2 types, 2 molecules AB
| #1: DNA chain | Mass: 2122.424 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) |
|---|---|
| #2: DNA chain | Mass: 2138.423 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) |
-Protein/peptide , 2 types, 2 molecules CD
-Non-polymers , 3 types, 28 molecules 
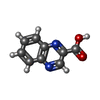



| #5: Chemical | | #6: Chemical |   Type: Cyclic depsipeptide / Class: Antibiotic / Mass: 174.156 Da / Num. of mol.: 2 / Source method: obtained synthetically / Formula: C9H6N2O2 Type: Cyclic depsipeptide / Class: Antibiotic / Mass: 174.156 Da / Num. of mol.: 2 / Source method: obtained synthetically / Formula: C9H6N2O2Details: ECHINOMYCIN IS A BICYCLIC OCTADEPSIPEPTIDE. BICYCLIZATION IS ACHIEVED BY LINKING THE N- AND THE C- TERMINI, AND A THIOACETAL BOND BETWEEN RESIDUES 3 AND 7. THE TWO QUINOXALINE CHROMOPHORES ...Details: ECHINOMYCIN IS A BICYCLIC OCTADEPSIPEPTIDE. BICYCLIZATION IS ACHIEVED BY LINKING THE N- AND THE C- TERMINI, AND A THIOACETAL BOND BETWEEN RESIDUES 3 AND 7. THE TWO QUINOXALINE CHROMOPHORES ARE LINKED TO THE D-SERINE RESIDUES, RESIDUES 1 AND 5. References: Echinomycin #7: Water | ChemComp-HOH / | |
|---|
-Details
| Compound details | ACTINOMYCIN D IS A BICYCLIC PEPTIDE, A MEMBER OF THE ACTINOMYCIN FAMILY. HERE, ACTINOMYCIN D IS ...ACTINOMYCI |
|---|---|
| Has ligand of interest | N |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.43 Å3/Da / Density % sol: 49.37 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 6.5 Details: 0.25 mM Oligonucleotide, 0.25 mM Echinomycin, 0.125 mM Actinomycin D, 2.5 mM Sodium cacodylate (pH 6.5), 2.5 mM magnesium chloride, 2 mM spermine tetrahydrochloride, 6 mM cobalt chloride, 2% ...Details: 0.25 mM Oligonucleotide, 0.25 mM Echinomycin, 0.125 mM Actinomycin D, 2.5 mM Sodium cacodylate (pH 6.5), 2.5 mM magnesium chloride, 2 mM spermine tetrahydrochloride, 6 mM cobalt chloride, 2% v/v PEG 200, 4% v/v MPD, Reservoir PEG 200 (30%) |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSRRC NSRRC  / Beamline: BL15A1 / Wavelength: 1 Å / Beamline: BL15A1 / Wavelength: 1 Å |
| Detector | Type: RAYONIX MX300HE / Detector: CCD / Date: Jan 11, 2018 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.89→30 Å / Num. obs: 5402 / % possible obs: 99.4 % / Redundancy: 13.3 % / Rmerge(I) obs: 0.04 / Rsym value: 0.04 / Net I/σ(I): 43.112 |
| Reflection shell | Resolution: 1.89→1.92 Å / Rmerge(I) obs: 0.343 / Mean I/σ(I) obs: 10.111 / Num. unique obs: 71669 / Rsym value: 0.343 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 7DQ0 Resolution: 1.89→27.723 Å / SU ML: 0.26 / Cross valid method: THROUGHOUT / σ(F): 1.35 / Phase error: 35.22 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 149.05 Å2 / Biso mean: 49.3143 Å2 / Biso min: 24.23 Å2 | ||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.89→27.723 Å
| ||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0
| ||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: 59.4546 Å / Origin y: 51.6124 Å / Origin z: 82.6273 Å
| ||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj