+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7nri | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the darobactin-bound E. coli BAM complex (BamABCDE) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Components Components |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | MEMBRANE PROTEIN / Beta-Barrel / outer membrane / protein insertion / protein folding / protein maturation / antibiotic / natural product / cyclized peptide | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationBam protein complex / Gram-negative-bacterium-type cell outer membrane assembly / Secretion of toxins / protein insertion into membrane / cell outer membrane / protein-macromolecule adaptor activity / cell adhesion / response to antibiotic / cell surface / identical protein binding / membrane Similarity search - Function | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological species |  synthetic construct (others) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Authors Authors | Jakob, R.P. / Kaur, H. / Marzinek, J.K. / Green, R. / Imai, Y. / Bolla, J. / Robinson, C. / Bond, P.J. / Lewis, K. / Maier, T. / Hiller, S. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: The antibiotic darobactin mimics a β-strand to inhibit outer membrane insertase. Authors: Hundeep Kaur / Roman P Jakob / Jan K Marzinek / Robert Green / Yu Imai / Jani Reddy Bolla / Elia Agustoni / Carol V Robinson / Peter J Bond / Kim Lewis / Timm Maier / Sebastian Hiller /     Abstract: Antibiotics that target Gram-negative bacteria in new ways are needed to resolve the antimicrobial resistance crisis. Gram-negative bacteria are protected by an additional outer membrane, rendering ...Antibiotics that target Gram-negative bacteria in new ways are needed to resolve the antimicrobial resistance crisis. Gram-negative bacteria are protected by an additional outer membrane, rendering proteins on the cell surface attractive drug targets. The natural compound darobactin targets the bacterial insertase BamA-the central unit of the essential BAM complex, which facilitates the folding and insertion of outer membrane proteins. BamA lacks a typical catalytic centre, and it is not obvious how a small molecule such as darobactin might inhibit its function. Here we resolve the mode of action of darobactin at the atomic level using a combination of cryo-electron microscopy, X-ray crystallography, native mass spectrometry, in vivo experiments and molecular dynamics simulations. Two cyclizations pre-organize the darobactin peptide in a rigid β-strand conformation. This creates a mimic of the recognition signal of native substrates with a superior ability to bind to the lateral gate of BamA. Upon binding, darobactin replaces a lipid molecule from the lateral gate to use the membrane environment as an extended binding pocket. Because the interaction between darobactin and BamA is largely mediated by backbone contacts, it is particularly robust against potential resistance mutations. Our results identify the lateral gate as a functional hotspot in BamA and will allow the rational design of antibiotics that target this bacterial Achilles heel. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7nri.cif.gz 7nri.cif.gz | 495.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7nri.ent.gz pdb7nri.ent.gz | 402.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7nri.json.gz 7nri.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/nr/7nri https://data.pdbj.org/pub/pdb/validation_reports/nr/7nri ftp://data.pdbj.org/pub/pdb/validation_reports/nr/7nri ftp://data.pdbj.org/pub/pdb/validation_reports/nr/7nri | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  12546MC 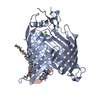 7nreC  7nrfC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Outer membrane protein assembly factor ... , 5 types, 5 molecules ABCDE
| #1: Protein | Mass: 88227.430 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Strain: K12 / Gene: bamA, yaeT, yzzN, yzzY, b0177, JW0172 / Plasmid: pQLink / Production host:  |
|---|---|
| #2: Protein | Mass: 40924.516 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Strain: K12 / Gene: bamB, yfgL, b2512, JW2496 / Plasmid: pQlink / Production host:  |
| #3: Protein | Mass: 34401.250 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Strain: K12 / Gene: bamC, dapX, nlpB, b2477, JW2462 / Plasmid: pQlink / Production host:  |
| #4: Protein | Mass: 25816.818 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Strain: K12 / Gene: bamD, yfiO, b2595, JW2577 / Plasmid: pQLink / Production host:  |
| #5: Protein | Mass: 11462.772 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Strain: K12 / Gene: bamE, smpA, b2617, JW2598 / Plasmid: pQLink / Production host:  |
-Protein/peptide , 1 types, 1 molecules G
| #6: Protein/peptide | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
| Nonpolymer details | Original Refinement was done with Darobactin defined as one ligand |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: BamABCDE complex / Type: COMPLEX / Entity ID: #2-#5 / Source: RECOMBINANT | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Experimental value: NO | ||||||||||||||||||||
| Source (natural) | Organism:  | ||||||||||||||||||||
| Source (recombinant) | Organism:  | ||||||||||||||||||||
| Buffer solution | pH: 8 | ||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||
| Specimen | Conc.: 5.5 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||
| Specimen support | Grid material: COPPER / Grid mesh size: 200 divisions/in. / Grid type: C-flat-1.2/1.3 | ||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK III / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 293 K / Details: 4microL of sample applied |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS / Details: Manually grid screening, operated by SerialEM |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 165000 X / Nominal defocus max: 1800 nm / Nominal defocus min: 800 nm / Calibrated defocus min: 500 nm / Calibrated defocus max: 2200 nm / Cs: 2.7 mm / C2 aperture diameter: 70 µm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 9 sec. / Electron dose: 50 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Num. of grids imaged: 1 / Num. of real images: 5527 |
| EM imaging optics | Energyfilter name: GIF Bioquantum / Energyfilter slit width: 20 eV |
| Image scans | Movie frames/image: 45 |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.17.1_3660: / Classification: refinement | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||
| CTF correction | Type: NONE | ||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 870023 | ||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.03 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 157474 / Num. of class averages: 1 / Symmetry type: POINT | ||||||||||||||||||||||||||||||
| Atomic model building | Protocol: AB INITIO MODEL / Space: REAL | ||||||||||||||||||||||||||||||
| Atomic model building | PDB-ID: 5D0O Accession code: 5D0O / Source name: PDB / Type: experimental model | ||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj