[English] 日本語
 Yorodumi
Yorodumi- PDB-7m32: Dihydropyrimidine Dehydrogenase (DPD) C671A Mutant Soaked with Ur... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7m32 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Dihydropyrimidine Dehydrogenase (DPD) C671A Mutant Soaked with Uracil and NADPH Anaerobically | ||||||
 Components Components | Dihydropyrimidine dehydrogenase [NADP(+)] | ||||||
 Keywords Keywords | FLAVOPROTEIN / Oxidoreductase / Dihydropyrimidine Dehydrogenase / DPD / mutant / uracil / flavin / soaking / ligand / FMN / FAD | ||||||
| Function / homology |  Function and homology information Function and homology informationdihydropyrimidine dehydrogenase (NADP+) / uracil binding / thymidine catabolic process / dihydropyrimidine dehydrogenase (NADP+) activity / beta-alanine biosynthetic process / uracil catabolic process / thymine catabolic process / FMN binding / NADP binding / flavin adenine dinucleotide binding ...dihydropyrimidine dehydrogenase (NADP+) / uracil binding / thymidine catabolic process / dihydropyrimidine dehydrogenase (NADP+) activity / beta-alanine biosynthetic process / uracil catabolic process / thymine catabolic process / FMN binding / NADP binding / flavin adenine dinucleotide binding / 4 iron, 4 sulfur cluster binding / protein homodimerization activity / metal ion binding / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.82 Å MOLECULAR REPLACEMENT / Resolution: 1.82 Å | ||||||
 Authors Authors | Butrin, A. / Beaupre, B. / Forouzesh, D. / Liu, D. / Moran, G. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2021 Journal: Biochemistry / Year: 2021Title: Perturbing the Movement of Hydrogens to Delineate and Assign Events in the Reductive Activation and Turnover of Porcine Dihydropyrimidine Dehydrogenase. Authors: Beaupre, B.A. / Forouzesh, D.C. / Butrin, A. / Liu, D. / Moran, G.R. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7m32.cif.gz 7m32.cif.gz | 846.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7m32.ent.gz pdb7m32.ent.gz | 680.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7m32.json.gz 7m32.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/m3/7m32 https://data.pdbj.org/pub/pdb/validation_reports/m3/7m32 ftp://data.pdbj.org/pub/pdb/validation_reports/m3/7m32 ftp://data.pdbj.org/pub/pdb/validation_reports/m3/7m32 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7m31C  1h7wS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 4 molecules ABCD
| #1: Protein | Mass: 111571.281 Da / Num. of mol.: 4 / Mutation: C671A Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: Q28943, dihydropyrimidine dehydrogenase (NADP+) |
|---|
-Non-polymers , 8 types, 2663 molecules 

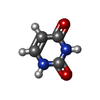











| #2: Chemical | ChemComp-SF4 / #3: Chemical | ChemComp-FAD / #4: Chemical | ChemComp-URA / #5: Chemical | ChemComp-FNR / #6: Chemical | #7: Chemical | #8: Chemical | ChemComp-PRO / | #9: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.36 Å3/Da / Density % sol: 47.8 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop Details: ~4.5 mg/mL (~40 uM) of C671A DPD in 25 mM HEPES, 2 mM DTT, 10% glycerol at pH 7.5 was prepared and diluted 1:1 with well solution containing 100 mM sodium citrate, 2 mM DTT, 15% PEG 6000 at ...Details: ~4.5 mg/mL (~40 uM) of C671A DPD in 25 mM HEPES, 2 mM DTT, 10% glycerol at pH 7.5 was prepared and diluted 1:1 with well solution containing 100 mM sodium citrate, 2 mM DTT, 15% PEG 6000 at pH 4.5 to 4.9. Diffraction quality crystals with elongated morphology were obtained by the hanging-drop vapor diffusion method. Incubation was carried out in the dark to eliminate photo-degradation the quasi-labile FMN cofactor. Crystals appeared after 16 hours as both single elongated rectangular hexahedron forms (200 x 50 x 50) or urchin-like clusters. Only the single crystal form were harvested and these were placed in a Plas-Labs 830 series glove box into which a Motic binocular microscope coupled to an Accuscope 1080 P HD camera was placed. Before being placed in the glove box, the well solutions of the selected crystals were made anaerobic with the addition of 10 mM dithionite and resealed with the cover slide. The glove box was then made anaerobic by flushing with pure nitrogen for approximately 10 minutes at which time the fractional dioxygen was 0.1 %, as indicated by a Forensics Detectors oxygen meter. Atmospheric dioxygen was measured throughout the soaking procedure and was held below 1%. C671A DPD crystals were soaked for a minimum of 20 minutes in 25 mM HEPES, 100 mM sodium citrate, 2 mM DTT, 100 uM NADPH, 100 uM uracil, 20% PEG 6000, 20% PEG 400, pH 7.5 prior to submersion in liquid nitrogen. Frozen crystals were then removed from the anaerobic environment and stored under liquid nitrogen. |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 21-ID-D / Wavelength: 1.1272 Å / Beamline: 21-ID-D / Wavelength: 1.1272 Å | ||||||||||||||||||||||||||||||
| Detector | Type: DECTRIS EIGER X 9M / Detector: PIXEL / Date: Dec 9, 2020 | ||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1.1272 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.82→51.69 Å / Num. obs: 354924 / % possible obs: 96.5 % / Redundancy: 4.8 % / Biso Wilson estimate: 25.22 Å2 / CC1/2: 0.996 / Rmerge(I) obs: 0.147 / Rpim(I) all: 0.074 / Rrim(I) all: 0.165 / Net I/σ(I): 7.4 / Num. measured all: 1705022 / Scaling rejects: 41 | ||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1H7W Resolution: 1.82→47.26 Å / SU ML: 0.24 / Cross valid method: THROUGHOUT / σ(F): 1.34 / Phase error: 21.29 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 131.41 Å2 / Biso mean: 34.5565 Å2 / Biso min: 13.88 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.82→47.26 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0 / Total num. of bins used: 30
|
 Movie
Movie Controller
Controller











 PDBj
PDBj




















