[English] 日本語
 Yorodumi
Yorodumi- PDB-7lml: Receptor for Advanced Glycation End Products VC1 domain in comple... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7lml | ||||||
|---|---|---|---|---|---|---|---|
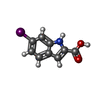
| Title | Receptor for Advanced Glycation End Products VC1 domain in complex with 3-(3-(((3-(4-Carboxyphenoxy)benzyl)oxy)methyl)phenyl)-1H-indole-2-carboxylic acid | ||||||
 Components Components | Advanced glycosylation end product-specific receptor | ||||||
 Keywords Keywords | SIGNALING PROTEIN / RAGE / IG-like domain / receptor / Advanced Glycation End Products | ||||||
| Function / homology |  Function and homology information Function and homology informationregulation of CD4-positive, alpha-beta T cell activation / negative regulation of blood circulation / positive regulation of endothelin production / advanced glycation end-product receptor activity / glucose mediated signaling pathway / positive regulation of monocyte extravasation / regulation of T cell mediated cytotoxicity / positive regulation of DNA-templated DNA replication / positive regulation of dendritic cell differentiation / negative regulation of long-term synaptic depression ...regulation of CD4-positive, alpha-beta T cell activation / negative regulation of blood circulation / positive regulation of endothelin production / advanced glycation end-product receptor activity / glucose mediated signaling pathway / positive regulation of monocyte extravasation / regulation of T cell mediated cytotoxicity / positive regulation of DNA-templated DNA replication / positive regulation of dendritic cell differentiation / negative regulation of long-term synaptic depression / regulation of p38MAPK cascade / regulation of non-canonical NF-kappaB signal transduction / positive regulation of amyloid precursor protein catabolic process / transcytosis / induction of positive chemotaxis / positive regulation of heterotypic cell-cell adhesion / S100 protein binding / positive regulation of monocyte chemotactic protein-1 production / positive regulation of p38MAPK cascade / regulation of long-term synaptic potentiation / protein localization to membrane / regulation of spontaneous synaptic transmission / negative regulation of connective tissue replacement involved in inflammatory response wound healing / scavenger receptor activity / negative regulation of interleukin-10 production / positive regulation of double-strand break repair / laminin receptor activity / response to amyloid-beta / TRAF6 mediated NF-kB activation / negative regulation of long-term synaptic potentiation / Advanced glycosylation endproduct receptor signaling / positive regulation of activated T cell proliferation / phagocytosis / transport across blood-brain barrier / positive regulation of chemokine production / phagocytic cup / positive regulation of interleukin-12 production / astrocyte activation / positive regulation of interleukin-1 beta production / microglial cell activation / positive regulation of non-canonical NF-kappaB signal transduction / : / TAK1-dependent IKK and NF-kappa-B activation / regulation of synaptic plasticity / positive regulation of JNK cascade / response to wounding / positive regulation of interleukin-6 production / cellular response to amyloid-beta / fibrillar center / neuron projection development / positive regulation of tumor necrosis factor production / transmembrane signaling receptor activity / cell junction / signaling receptor activity / amyloid-beta binding / regulation of inflammatory response / histone binding / molecular adaptor activity / response to hypoxia / learning or memory / early endosome / cell surface receptor signaling pathway / positive regulation of ERK1 and ERK2 cascade / apical plasma membrane / postsynapse / inflammatory response / protein-containing complex binding / cell surface / DNA binding / RNA binding / extracellular region / identical protein binding / nucleus / plasma membrane Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.15 Å MOLECULAR REPLACEMENT / Resolution: 2.15 Å | ||||||
 Authors Authors | Salay, L.E. / Kozlyuk, N. / Gilston, B.A. / Gogliotti, R.D. / Christov, P.P. / Kim, K. / Ovee, M. / Waterson, A.G. / Chazin, W.J. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Proteins / Year: 2021 Journal: Proteins / Year: 2021Title: A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors. Authors: Kozlyuk, N. / Gilston, B.A. / Salay, L.E. / Gogliotti, R.D. / Christov, P.P. / Kim, K. / Ovee, M. / Waterson, A.G. / Chazin, W.J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7lml.cif.gz 7lml.cif.gz | 214 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7lml.ent.gz pdb7lml.ent.gz | 142.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7lml.json.gz 7lml.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/lm/7lml https://data.pdbj.org/pub/pdb/validation_reports/lm/7lml ftp://data.pdbj.org/pub/pdb/validation_reports/lm/7lml ftp://data.pdbj.org/pub/pdb/validation_reports/lm/7lml | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6xq1C  6xq3C  6xq5C  6xq6C  6xq7C  6xq8C  6xq9C  7lmwC  4lp4S C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||||||||
| Unit cell |
| ||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Component-ID: 1 / Ens-ID: ens_1 / Beg auth comp-ID: GLY / Beg label comp-ID: GLY / End auth comp-ID: GLU / End label comp-ID: GLU / Auth seq-ID: 20 - 231 / Label seq-ID: 1 - 212
NCS oper: (Code: givenMatrix: (0.501489472128, -0.865162905542, 0.00120673901847), (-0.865163245968, -0.501490195115, -0.00037686831584), (0.000931220272965, -0.000855030753486, -0.999999200875) ...NCS oper: (Code: given Matrix: (0.501489472128, -0.865162905542, 0.00120673901847), Vector: |
- Components
Components
| #1: Protein | Mass: 23131.494 Da / Num. of mol.: 2 / Fragment: VC1 domain, resdues 23-231 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: AGER, RAGE / Production host: Homo sapiens (human) / Gene: AGER, RAGE / Production host:  #2: Chemical | ChemComp-ACT / #3: Chemical | #4: Chemical | ChemComp-Y6S / #5: Water | ChemComp-HOH / | Has ligand of interest | N | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.35 Å3/Da / Density % sol: 63.25 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 7.5 / Details: 2.5 M NaOAc (pH 7.5) |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 12.3.1 / Wavelength: 0.9795 Å / Beamline: 12.3.1 / Wavelength: 0.9795 Å |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: May 12, 2017 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9795 Å / Relative weight: 1 |
| Reflection | Resolution: 2.14983→40.7005 Å / Num. obs: 32950 / % possible obs: 99.3 % / Redundancy: 4 % / Biso Wilson estimate: 25.2 Å2 / Rpim(I) all: 0.026 / Rrim(I) all: 0.072 / Net I/σ(I): 16.2 |
| Reflection shell | Resolution: 2.14983→2.5319 Å / Redundancy: 4 % / Mean I/σ(I) obs: 3.66 / Num. unique obs: 12707 / Rpim(I) all: 0.044 / Rrim(I) all: 0.124 / % possible all: 99.1 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4LP4 Resolution: 2.15→40.7 Å / SU ML: 0.2046 / Cross valid method: FREE R-VALUE / σ(F): 1.37 / Phase error: 25.2278 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 34.81 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.15→40.7 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | Type: Torsion NCS / Rms dev position: 0.383018016412 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: 30.9819022704 Å / Origin y: -17.8654547131 Å / Origin z: -14.2154855279 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group | Selection details: all |
 Movie
Movie Controller
Controller












 PDBj
PDBj