Entry Database : PDB / ID : 7jxnTitle Beta hairpin derived from Abeta17-36 with an F20Cha mutation Amyloid-beta 17-36 peptide Keywords / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / / Resolution : 2 Å Authors Kreutzer, A.G. / Haerianardakani, S. / Nowick, J.S. Funding support Organization Grant number Country National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) GM097562
Journal : J.Am.Chem.Soc. / Year : 2020Title : Phenylalanine Mutation to Cyclohexylalanine Facilitates Triangular Trimer Formation by beta-Hairpins Derived from A beta.Authors : Haerianardakani, S. / Kreutzer, A.G. / Salveson, P.J. / Samdin, T.D. / Guaglianone, G.E. / Nowick, J.S. History Deposition Aug 27, 2020 Deposition site / Processing site Revision 1.0 Dec 9, 2020 Provider / Type Revision 1.1 Dec 23, 2020 Group / Category / citation_authorItem _citation.journal_volume / _citation.page_first ... _citation.journal_volume / _citation.page_first / _citation.page_last / _citation_author.identifier_ORCID Revision 1.2 Apr 2, 2025 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Structure summary Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_entry_details / pdbx_modification_feature / struct_conn Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_entry_details.has_protein_modification / _struct_conn.pdbx_leaving_atom_flag
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  SAD / Resolution: 2 Å
SAD / Resolution: 2 Å  Authors
Authors United States, 1items
United States, 1items  Citation
Citation Journal: J.Am.Chem.Soc. / Year: 2020
Journal: J.Am.Chem.Soc. / Year: 2020 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 7jxn.cif.gz
7jxn.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb7jxn.ent.gz
pdb7jxn.ent.gz PDB format
PDB format 7jxn.json.gz
7jxn.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads 7jxn_validation.pdf.gz
7jxn_validation.pdf.gz wwPDB validaton report
wwPDB validaton report 7jxn_full_validation.pdf.gz
7jxn_full_validation.pdf.gz 7jxn_validation.xml.gz
7jxn_validation.xml.gz 7jxn_validation.cif.gz
7jxn_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/jx/7jxn
https://data.pdbj.org/pub/pdb/validation_reports/jx/7jxn ftp://data.pdbj.org/pub/pdb/validation_reports/jx/7jxn
ftp://data.pdbj.org/pub/pdb/validation_reports/jx/7jxn Links
Links Assembly
Assembly
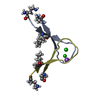

 Components
Components Homo sapiens (human) / References: UniProt: P05067
Homo sapiens (human) / References: UniProt: P05067 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  ALS
ALS  / Beamline: 8.2.2 / Wavelength: 0.98 Å
/ Beamline: 8.2.2 / Wavelength: 0.98 Å Processing
Processing SAD / Resolution: 2→32.19 Å / Cross valid method: THROUGHOUT / σ(F): 14.73 / Phase error: 33.22 / Stereochemistry target values: TWIN_LSQ_F
SAD / Resolution: 2→32.19 Å / Cross valid method: THROUGHOUT / σ(F): 14.73 / Phase error: 33.22 / Stereochemistry target values: TWIN_LSQ_F Movie
Movie Controller
Controller




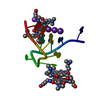







 PDBj
PDBj




















