[English] 日本語
 Yorodumi
Yorodumi- PDB-7ckq: The cryo-EM structure of B. subtilis BmrR transcription activatio... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7ckq | ||||||
|---|---|---|---|---|---|---|---|
| Title | The cryo-EM structure of B. subtilis BmrR transcription activation complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSCRIPTION / RNA polymerase / Transcription activation | ||||||
| Function / homology |  Function and homology information Function and homology informationnucleoid / sigma factor activity / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / DNA-directed RNA polymerase complex / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / protein dimerization activity ...nucleoid / sigma factor activity / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / DNA-directed RNA polymerase complex / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / protein dimerization activity / DNA-binding transcription factor activity / response to antibiotic / regulation of DNA-templated transcription / DNA-templated transcription / magnesium ion binding / DNA binding / zinc ion binding / cytoplasm Similarity search - Function | ||||||
| Biological species |  synthetic construct (others) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 4.4 Å | ||||||
 Authors Authors | Fang, C.L. / Zhang, Y. | ||||||
| Funding support |  China, 1items China, 1items
| ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: The bacterial multidrug resistance regulator BmrR distorts promoter DNA to activate transcription. Authors: Chengli Fang / Linyu Li / Yihan Zhao / Xiaoxian Wu / Steven J Philips / Linlin You / Mingkang Zhong / Xiaojin Shi / Thomas V O'Halloran / Qunyi Li / Yu Zhang /   Abstract: The MerR-family proteins represent a unique family of bacteria transcription factors (TFs), which activate transcription in a manner distinct from canonical ones. Here, we report a cryo-EM structure ...The MerR-family proteins represent a unique family of bacteria transcription factors (TFs), which activate transcription in a manner distinct from canonical ones. Here, we report a cryo-EM structure of a B. subtilis transcription activation complex comprising B. subtilis six-subunit (2αββ'ωε) RNA Polymerase (RNAP) core enzyme, σ, a promoter DNA, and the ligand-bound B. subtilis BmrR, a prototype of MerR-family TFs. The structure reveals that RNAP and BmrR recognize the upstream promoter DNA from opposite faces and induce four significant kinks from the -35 element to the -10 element of the promoter DNA in a cooperative manner, which restores otherwise inactive promoter activity by shortening the length of promoter non-optimal -35/-10 spacer. Our structure supports a DNA-distortion and RNAP-non-contact paradigm of transcriptional activation by MerR TFs. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7ckq.cif.gz 7ckq.cif.gz | 654 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7ckq.ent.gz pdb7ckq.ent.gz | 504.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7ckq.json.gz 7ckq.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ck/7ckq https://data.pdbj.org/pub/pdb/validation_reports/ck/7ckq ftp://data.pdbj.org/pub/pdb/validation_reports/ck/7ckq ftp://data.pdbj.org/pub/pdb/validation_reports/ck/7ckq | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  30390MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-DNA-directed RNA polymerase subunit ... , 4 types, 5 molecules ABCDE
| #1: Protein | Mass: 34842.387 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #2: Protein | | Mass: 133847.938 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  #3: Protein | | Mass: 134444.953 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  #4: Protein | | Mass: 7766.921 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|
-Protein , 3 types, 4 molecules FGIH
| #5: Protein | Mass: 43007.406 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  | ||
|---|---|---|---|
| #8: Protein | Mass: 32951.688 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Strain: 168 / Gene: bmrR, bmr1R, BSU24020 / Production host:  #9: Protein | | Mass: 8263.358 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-DNA chain , 2 types, 2 molecules 12
| #6: DNA chain | Mass: 15489.907 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) |
|---|---|
| #7: DNA chain | Mass: 15485.940 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) |
-Non-polymers , 3 types, 5 molecules 

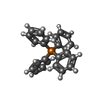


| #10: Chemical | ChemComp-MG / | ||
|---|---|---|---|
| #11: Chemical | | #12: Chemical | |
-Details
| Has ligand of interest | N |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Bacillus subtilis BmrR transcription activation complex Type: COMPLEX / Entity ID: #1-#9 / Source: RECOMBINANT | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.48 MDa / Experimental value: NO | |||||||||||||||||||||||||
| Source (natural) | Organism:  | |||||||||||||||||||||||||
| Source (recombinant) | Organism:  | |||||||||||||||||||||||||
| Buffer solution | pH: 7.5 | |||||||||||||||||||||||||
| Buffer component |
| |||||||||||||||||||||||||
| Specimen | Conc.: 19 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | |||||||||||||||||||||||||
| Specimen support | Grid material: COPPER / Grid mesh size: 400 divisions/in. / Grid type: C-flat-1.2/1.3 | |||||||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 95 % / Chamber temperature: 282.65 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Cs: 2.7 mm |
| Image recording | Average exposure time: 7.6 sec. / Electron dose: 60.8 e/Å2 / Detector mode: SUPER-RESOLUTION / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Num. of grids imaged: 1 / Num. of real images: 2226 |
| Image scans | Movie frames/image: 38 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: NONE | ||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 494295 | ||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 4.4 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 103226 / Num. of class averages: 1 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT | ||||||||||||||||||||||||||||||||||||
| Atomic model building |
|
 Movie
Movie Controller
Controller









 PDBj
PDBj













































