[English] 日本語
 Yorodumi
Yorodumi- EMDB-30390: The cryo-EM structure of B. subtilis BmrR transcription activatio... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30390 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The cryo-EM structure of B. subtilis BmrR transcription activation complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA polymerase / Transcription activation / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationnucleoid / sigma factor activity / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / DNA-directed RNA polymerase complex / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / protein dimerization activity ...nucleoid / sigma factor activity / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / DNA-directed RNA polymerase complex / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / protein dimerization activity / DNA-binding transcription factor activity / response to antibiotic / DNA-templated transcription / regulation of DNA-templated transcription / magnesium ion binding / DNA binding / zinc ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
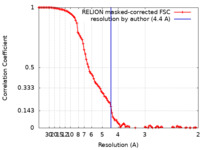
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Fang CL / Zhang Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: The bacterial multidrug resistance regulator BmrR distorts promoter DNA to activate transcription. Authors: Chengli Fang / Linyu Li / Yihan Zhao / Xiaoxian Wu / Steven J Philips / Linlin You / Mingkang Zhong / Xiaojin Shi / Thomas V O'Halloran / Qunyi Li / Yu Zhang /   Abstract: The MerR-family proteins represent a unique family of bacteria transcription factors (TFs), which activate transcription in a manner distinct from canonical ones. Here, we report a cryo-EM structure ...The MerR-family proteins represent a unique family of bacteria transcription factors (TFs), which activate transcription in a manner distinct from canonical ones. Here, we report a cryo-EM structure of a B. subtilis transcription activation complex comprising B. subtilis six-subunit (2αββ'ωε) RNA Polymerase (RNAP) core enzyme, σ, a promoter DNA, and the ligand-bound B. subtilis BmrR, a prototype of MerR-family TFs. The structure reveals that RNAP and BmrR recognize the upstream promoter DNA from opposite faces and induce four significant kinks from the -35 element to the -10 element of the promoter DNA in a cooperative manner, which restores otherwise inactive promoter activity by shortening the length of promoter non-optimal -35/-10 spacer. Our structure supports a DNA-distortion and RNAP-non-contact paradigm of transcriptional activation by MerR TFs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30390.map.gz emd_30390.map.gz | 9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30390-v30.xml emd-30390-v30.xml emd-30390.xml emd-30390.xml | 25.9 KB 25.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30390_fsc.xml emd_30390_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_30390.png emd_30390.png | 93.2 KB | ||
| Filedesc metadata |  emd-30390.cif.gz emd-30390.cif.gz | 9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30390 http://ftp.pdbj.org/pub/emdb/structures/EMD-30390 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30390 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30390 | HTTPS FTP |
-Related structure data
| Related structure data |  7ckqMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30390.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30390.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Bacillus subtilis BmrR transcription activation complex
+Supramolecule #1: Bacillus subtilis BmrR transcription activation complex
+Macromolecule #1: DNA-directed RNA polymerase subunit alpha
+Macromolecule #2: DNA-directed RNA polymerase subunit beta
+Macromolecule #3: DNA-directed RNA polymerase subunit beta'
+Macromolecule #4: DNA-directed RNA polymerase subunit omega
+Macromolecule #5: RNA polymerase sigma factor SigA
+Macromolecule #8: Multidrug-efflux transporter 1 regulator
+Macromolecule #9: UPF0356 protein YkzG
+Macromolecule #6: DNA (50-MER)
+Macromolecule #7: DNA (50-MER)
+Macromolecule #10: MAGNESIUM ION
+Macromolecule #11: ZINC ION
+Macromolecule #12: TETRAPHENYLPHOSPHONIUM
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 19 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 120 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: OTHER | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 282.65 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Number real images: 2226 / Average exposure time: 7.6 sec. / Average electron dose: 60.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Refinement | Protocol: RIGID BODY FIT | ||||||||
| Output model |  PDB-7ckq: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)