| Entry | Database: PDB / ID: 7ai3
|
|---|
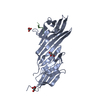
| Title | Crystal structure of MCE domain of Mce4A from Mycobacterium tuberculosis H37Rv |
|---|
 Components Components | Mce-family protein Mce4A |
|---|
 Keywords Keywords | TRANSPORT PROTEIN / substrate binding protein / MCE / Mce4A |
|---|
| Function / homology |  Function and homology information Function and homology information
phospholipid transfer activity / intermembrane phospholipid transfer / symbiont entry into host / peptidoglycan-based cell wall / plasma membraneSimilarity search - Function Virulence factor Mce protein / Mammalian cell entry, C-terminal / Cholesterol uptake porter CUP1 of Mce4, putative / : / Mce/MlaD / MlaD proteinSimilarity search - Domain/homology |
|---|
| Biological species |  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.9 Å MOLECULAR REPLACEMENT / Resolution: 2.9 Å |
|---|
 Authors Authors | Asthana, P. / Venkatesan, R. |
|---|
| Funding support |  Finland, 6items Finland, 6items | Organization | Grant number | Country |
|---|
| H2020 Marie Curie Actions of the European Commission | 713606 |  Finland Finland | | Jane and Aatos Erkko Foundation | |  Finland Finland | | Sigrid Juselius Foundation | |  Finland Finland | | European Union (EU) | 653706 |  Finland Finland | | Academy of Finland | 319194 |  Finland Finland | | European Commission | 730872 |  Finland Finland |
|
|---|
 Citation Citation |  Journal: Iucrj / Year: 2021 Journal: Iucrj / Year: 2021
Title: Structural insights into the substrate-binding proteins Mce1A and Mce4A from Mycobacterium tuberculosis .
Authors: Asthana, P. / Singh, D. / Pedersen, J.S. / Hynonen, M.J. / Sulu, R. / Murthy, A.V. / Laitaoja, M. / Janis, J. / Riley, L.W. / Venkatesan, R. |
|---|
| History | | Deposition | Sep 25, 2020 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | Aug 25, 2021 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Oct 13, 2021 | Group: Data collection / Database references / Source and taxonomy
Category: citation / citation_author ...citation / citation_author / database_PDB_rev / database_PDB_rev_record / entity_src_gen / pdbx_database_proc / pdbx_struct_ref_seq_depositor_info
Item: _citation.journal_volume / _citation.page_first ..._citation.journal_volume / _citation.page_first / _citation.page_last / _citation.pdbx_database_id_PubMed / _citation.title / _citation_author.identifier_ORCID / _entity_src_gen.gene_src_strain / _entity_src_gen.pdbx_gene_src_scientific_name / _pdbx_struct_ref_seq_depositor_info.db_seq_one_letter_code |
|---|
| Revision 1.2 | May 1, 2024 | Group: Data collection / Refinement description
Category: chem_comp_atom / chem_comp_bond / pdbx_initial_refinement_model |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Mycobacterium tuberculosis H37Rv (bacteria)
Mycobacterium tuberculosis H37Rv (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.9 Å
MOLECULAR REPLACEMENT / Resolution: 2.9 Å  Authors
Authors Finland, 6items
Finland, 6items  Citation
Citation Journal: Iucrj / Year: 2021
Journal: Iucrj / Year: 2021 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 7ai3.cif.gz
7ai3.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb7ai3.ent.gz
pdb7ai3.ent.gz PDB format
PDB format 7ai3.json.gz
7ai3.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/ai/7ai3
https://data.pdbj.org/pub/pdb/validation_reports/ai/7ai3 ftp://data.pdbj.org/pub/pdb/validation_reports/ai/7ai3
ftp://data.pdbj.org/pub/pdb/validation_reports/ai/7ai3 Links
Links Assembly
Assembly


 Components
Components Mycobacterium tuberculosis H37Rv (bacteria)
Mycobacterium tuberculosis H37Rv (bacteria)
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  MAX IV
MAX IV  / Beamline: BioMAX / Wavelength: 0.953 Å
/ Beamline: BioMAX / Wavelength: 0.953 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj