[English] 日本語
 Yorodumi
Yorodumi- PDB-6zbv: Inward-open structure of human glycine transporter 1 in complex w... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6zbv | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Inward-open structure of human glycine transporter 1 in complex with a benzoylisoindoline inhibitor and sybody Sb_GlyT1#7 | |||||||||||||||
 Components Components |
| |||||||||||||||
 Keywords Keywords | MEMBRANE PROTEIN / secondary active transport / neurotransmitter-sodium symport / amino acid transport / SLC6A9 / inward open state / inhibitor bound complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of synaptic transmission, glycinergic / glycine secretion, neurotransmission / glycine transmembrane transporter activity / glycine:sodium symporter activity / positive regulation of heme biosynthetic process / neurotransmitter uptake / glycine import across plasma membrane / glycine transport / positive regulation of hemoglobin biosynthetic process / amino acid:sodium symporter activity ...regulation of synaptic transmission, glycinergic / glycine secretion, neurotransmission / glycine transmembrane transporter activity / glycine:sodium symporter activity / positive regulation of heme biosynthetic process / neurotransmitter uptake / glycine import across plasma membrane / glycine transport / positive regulation of hemoglobin biosynthetic process / amino acid:sodium symporter activity / dense core granule / SLC-mediated transport of neurotransmitters / parallel fiber to Purkinje cell synapse / lateral plasma membrane / transport across blood-brain barrier / basal plasma membrane / sodium ion transmembrane transport / hippocampal mossy fiber to CA3 synapse / synaptic vesicle membrane / presynaptic membrane / basolateral plasma membrane / postsynaptic membrane / endosome / postsynaptic density / apical plasma membrane / membrane / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human)unidentified (others) | |||||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.4 Å MOLECULAR REPLACEMENT / Resolution: 3.4 Å | |||||||||||||||
 Authors Authors | Shahsavar, A. / Stohler, P. / Bourenkov, G. / Zimmermann, I. / Siegrist, M. / Guba, W. / Pinard, E. / Sinning, S. / Seeger, M.A. / Schneider, T.R. ...Shahsavar, A. / Stohler, P. / Bourenkov, G. / Zimmermann, I. / Siegrist, M. / Guba, W. / Pinard, E. / Sinning, S. / Seeger, M.A. / Schneider, T.R. / Dawson, R.J.P. / Nissen, P. | |||||||||||||||
| Funding support |  Denmark, Denmark,  Switzerland, Switzerland,  Germany, 4items Germany, 4items
| |||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Structural insights into the inhibition of glycine reuptake. Authors: Shahsavar, A. / Stohler, P. / Bourenkov, G. / Zimmermann, I. / Siegrist, M. / Guba, W. / Pinard, E. / Sinning, S. / Seeger, M.A. / Schneider, T.R. / Dawson, R.J.P. / Nissen, P. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6zbv.cif.gz 6zbv.cif.gz | 255.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6zbv.ent.gz pdb6zbv.ent.gz | 205.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6zbv.json.gz 6zbv.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/zb/6zbv https://data.pdbj.org/pub/pdb/validation_reports/zb/6zbv ftp://data.pdbj.org/pub/pdb/validation_reports/zb/6zbv ftp://data.pdbj.org/pub/pdb/validation_reports/zb/6zbv | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6zplC  4us3S  6dzzS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 64801.488 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: SLC6A9 / Plasmid: pCDNA3.1 / Cell (production host): HEK293 Homo sapiens (human) / Gene: SLC6A9 / Plasmid: pCDNA3.1 / Cell (production host): HEK293Production host: Mammalian expression vector EGFP-MCS-pcDNA3.1 (others) References: UniProt: P48067 |
|---|---|
| #2: Antibody | Mass: 13303.686 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.) unidentified (others) / Production host:  |
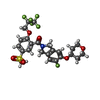
| #3: Chemical | ChemComp-QET / [ |
| Has ligand of interest | Y |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.92 Å3/Da / Density % sol: 57.89 % Description: Crystals appeared in 3-10 days with the longest dimension of 2-5 um. |
|---|---|
| Crystal grow | Temperature: 292.75 K / Method: lipidic cubic phase / pH: 7 Details: 0.1M ADA pH 7, 13-25% PEG600, 4-14% v/v-1,3-Butanediol |
-Data collection
| Diffraction |
| |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source |
| |||||||||||||||||||||||||||||||||||||||||||||
| Detector |
| |||||||||||||||||||||||||||||||||||||||||||||
| Radiation |
| |||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength |
| |||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 3.4→30 Å / Num. obs: 12641 / % possible obs: 99.71 % / Redundancy: 12.1 % / Biso Wilson estimate: 103.12 Å2 / CC1/2: 0.985 / CC star: 0.997 / Rmerge(I) obs: 0.42 / Rpim(I) all: 0.1225 / Rrim(I) all: 0.44 / Net I/σ(I): 6.57 | |||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Resolution: 3.4→3.5 Å / Redundancy: 11.9 % / Rmerge(I) obs: 2.688 / Mean I/σ(I) obs: 1.69 / Num. unique obs: 1232 / CC1/2: 0.541 / CC star: 0.838 / Rpim(I) all: 0.7956 / Rrim(I) all: 2.809 / % possible all: 100 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4us3, 6dzz Resolution: 3.4→29.67 Å / Cor.coef. Fo:Fc: 0.908 / Cor.coef. Fo:Fc free: 0.861 / Cross valid method: THROUGHOUT / σ(F): 0 / SU Rfree Blow DPI: 0.547
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 196.18 Å2 / Biso mean: 113.5 Å2 / Biso min: 77.2 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 3.4→29.67 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 3.4→3.44 Å / Rfactor Rfree error: 0 / Total num. of bins used: 32
|
 Movie
Movie Controller
Controller












 PDBj
PDBj