Entry Database : PDB / ID : 6yhpTitle Solution NMR Structure of APP V44M mutant TMD Amyloid-beta precursor protein V44M mutant Keywords / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / Authors Silber, M. / Muhle-Goll, C. Funding support Organization Grant number Country German Research Foundation (DFG) FOR2290
Journal : Acs Chem Neurosci / Year : 2020Title : Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.Authors : Silber, M. / Hitzenberger, M. / Zacharias, M. / Muhle-Goll, C. History Deposition Mar 30, 2020 Deposition site / Processing site Revision 1.0 Dec 9, 2020 Provider / Type Revision 1.1 Dec 23, 2020 Group / Category / citation_authorItem _citation.journal_volume / _citation.page_first ... _citation.journal_volume / _citation.page_first / _citation.page_last / _citation_author.identifier_ORCID Revision 1.2 May 15, 2024 Group / Database references / Category / chem_comp_bond / database_2Item / _database_2.pdbx_database_accession
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) Authors
Authors Germany, 1items
Germany, 1items  Citation
Citation Journal: Acs Chem Neurosci / Year: 2020
Journal: Acs Chem Neurosci / Year: 2020 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6yhp.cif.gz
6yhp.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6yhp.ent.gz
pdb6yhp.ent.gz PDB format
PDB format 6yhp.json.gz
6yhp.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/yh/6yhp
https://data.pdbj.org/pub/pdb/validation_reports/yh/6yhp ftp://data.pdbj.org/pub/pdb/validation_reports/yh/6yhp
ftp://data.pdbj.org/pub/pdb/validation_reports/yh/6yhp



 Links
Links Assembly
Assembly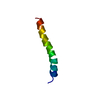
 Components
Components Homo sapiens (human) / References: UniProt: P05067
Homo sapiens (human) / References: UniProt: P05067 Sample preparation
Sample preparation Processing
Processing Movie
Movie Controller
Controller








 PDBj
PDBj














