[English] 日本語
 Yorodumi
Yorodumi- PDB-1r7f: NMR structure of the membrane anchor domain (1-31) of the nonstru... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1r7f | ||||||
|---|---|---|---|---|---|---|---|
| Title | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Ensemble of 43 structures. Sample in 100mM SDS) | ||||||
 Components Components | Genome polyprotein | ||||||
 Keywords Keywords | MEMBRANE PROTEIN / Membrane anchor domain / HCV NS5A protein / peptide. | ||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of hexokinase activity / translocation of peptides or proteins into host cell cytoplasm / symbiont-mediated perturbation of host cellular process / Toll-like receptor 2 binding / viral capsid assembly / adhesion receptor-mediated virion attachment to host cell / hepacivirin / TBC/RABGAPs / host cell mitochondrial membrane / host cell lipid droplet ...positive regulation of hexokinase activity / translocation of peptides or proteins into host cell cytoplasm / symbiont-mediated perturbation of host cellular process / Toll-like receptor 2 binding / viral capsid assembly / adhesion receptor-mediated virion attachment to host cell / hepacivirin / TBC/RABGAPs / host cell mitochondrial membrane / host cell lipid droplet / symbiont-mediated transformation of host cell / symbiont-mediated suppression of host TRAF-mediated signal transduction / positive regulation of cytokinesis / symbiont-mediated perturbation of host cell cycle G1/S transition checkpoint / negative regulation of protein secretion / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / endoplasmic reticulum-Golgi intermediate compartment membrane / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / SH3 domain binding / kinase binding / nucleoside-triphosphate phosphatase / channel activity / viral nucleocapsid / monoatomic ion transmembrane transport / clathrin-dependent endocytosis of virus by host cell / entry receptor-mediated virion attachment to host cell / RNA helicase activity / Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases / host cell perinuclear region of cytoplasm / host cell endoplasmic reticulum membrane / RNA helicase / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / ribonucleoprotein complex / serine-type endopeptidase activity / symbiont-mediated activation of host autophagy / viral translational frameshifting / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / fusion of virus membrane with host endosome membrane / viral envelope / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / negative regulation of transcription by RNA polymerase II / ATP hydrolysis activity / proteolysis / RNA binding / zinc ion binding / ATP binding Similarity search - Function | ||||||
| Method | SOLUTION NMR / distance geometry, simulated annealing, molecular dynamics, energy minimization | ||||||
 Authors Authors | Penin, F. / Brass, V. / Appel, N. / Ramboarina, S. / Montserret, R. / Ficheux, D. / Blum, H.E. / Bartenschlager, R. / Moradpour, D. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2004 Journal: J.Biol.Chem. / Year: 2004Title: Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. Authors: Penin, F. / Brass, V. / Appel, N. / Ramboarina, S. / Montserret, R. / Ficheux, D. / Blum, H.E. / Bartenschlager, R. / Moradpour, D. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1r7f.cif.gz 1r7f.cif.gz | 441.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1r7f.ent.gz pdb1r7f.ent.gz | 368.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1r7f.json.gz 1r7f.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/r7/1r7f https://data.pdbj.org/pub/pdb/validation_reports/r7/1r7f ftp://data.pdbj.org/pub/pdb/validation_reports/r7/1r7f ftp://data.pdbj.org/pub/pdb/validation_reports/r7/1r7f | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 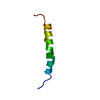
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 3770.423 Da / Num. of mol.: 1 Fragment: Nonstructural protein NS5A (P56)(residues 1973-2003 of Swiss-Prot sequence P27958) Source method: obtained synthetically Details: The peptide was chemically synthesized. The sequence is naturally found in hepatitis C virus. References: UniProt: P27958 |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 1.2mM NS5A[1-31], 10mM DTTd10 / Solvent system: 100mM SDS in H2O/D2O 95/5 (v/v) |
|---|---|
| Sample conditions | pH: 6 / Pressure: ambient / Temperature: 313 K |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| NMR spectrometer | Type: Varian UNITYPLUS / Manufacturer: Varian / Model: UNITYPLUS / Field strength: 500 MHz |
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: distance geometry, simulated annealing, molecular dynamics, energy minimization Software ordinal: 1 | ||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the least restraint violations Conformers calculated total number: 100 / Conformers submitted total number: 43 |
 Movie
Movie Controller
Controller












 PDBj
PDBj

 HSQC
HSQC