[English] 日本語
 Yorodumi
Yorodumi- PDB-6xok: X-ray structure of the rhombohedral form of the lipase from Therm... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6xok | ||||||
|---|---|---|---|---|---|---|---|
| Title | X-ray structure of the rhombohedral form of the lipase from Thermomyces lanuginosa at 1.3 A resolution | ||||||
 Components Components | Lipase | ||||||
 Keywords Keywords | LIPID BINDING PROTEIN / diacylglyceride / interfacial activation / oligomer / substrate complex | ||||||
| Function / homology |  Function and homology information Function and homology informationtriacylglycerol lipase / triacylglycerol lipase activity / lipid catabolic process Similarity search - Function | ||||||
| Biological species |   Thermomyces lanuginosus (fungus) Thermomyces lanuginosus (fungus) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.3 Å MOLECULAR REPLACEMENT / Resolution: 1.3 Å | ||||||
 Authors Authors | McPherson, A. | ||||||
 Citation Citation |  Journal: Current Enzyme Inhibition / Year: 2020 Journal: Current Enzyme Inhibition / Year: 2020Title: The crystal Structures of Thermomyces (Humicola) lanuginosa lipase in complex with enzymatic reactants Authors: McPherson, A. / Larson, S.B. / Kalasky, A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6xok.cif.gz 6xok.cif.gz | 229.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6xok.ent.gz pdb6xok.ent.gz | 154.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6xok.json.gz 6xok.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/xo/6xok https://data.pdbj.org/pub/pdb/validation_reports/xo/6xok ftp://data.pdbj.org/pub/pdb/validation_reports/xo/6xok ftp://data.pdbj.org/pub/pdb/validation_reports/xo/6xok | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6or3C  6xrvC  6xs3C  4ea6S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||||||||||||||
| 2 | x 6
| ||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||
| Components on special symmetry positions |
|
- Components
Components
-Protein / Sugars , 2 types, 2 molecules A

| #1: Protein | Mass: 31836.459 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Thermomyces lanuginosus (fungus) / Gene: LIP / Production host: Thermomyces lanuginosus (fungus) / Gene: LIP / Production host:  |
|---|---|
| #2: Sugar | ChemComp-NAG / |
-Non-polymers , 4 types, 435 molecules 





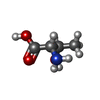
| #3: Chemical | ChemComp-PG4 / | ||||
|---|---|---|---|---|---|
| #4: Chemical | ChemComp-PO4 / #5: Chemical | ChemComp-LTV / | #6: Water | ChemComp-HOH / | |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.13 Å3/Da / Density % sol: 42.23 % / Description: thin needles of indeterminate cross section |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, sitting drop / pH: 6.5 Details: Crystals were grown by vapor diffusion at room temperature in 24 well Cryschem sitting drop plates (Hampton Research, Aliso Viejo, CA) with 600 microleters reservoirs of 25% PEG 3350 in 0.10 ...Details: Crystals were grown by vapor diffusion at room temperature in 24 well Cryschem sitting drop plates (Hampton Research, Aliso Viejo, CA) with 600 microleters reservoirs of 25% PEG 3350 in 0.10 M MES buffer at pH 6.5. The protein droplets were of 8ul volume and consisted of equal parts of a 20 to 30 mg/ml protein stock solution and the reservoir solution. Crystals usually appeared and grew to full size within one to two weeks. Rhombohedral crystals were thin needles of indeterminate cross-section PH range: 6.0 - 7.0 |
-Data collection
| Diffraction | Mean temperature: 173 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 8.3.1 / Wavelength: 1.03 Å / Beamline: 8.3.1 / Wavelength: 1.03 Å |
| Detector | Type: DECTRIS PILATUS3 X CdTe 1M / Detector: PIXEL / Date: Feb 20, 2019 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.03 Å / Relative weight: 1 |
| Reflection | Resolution: 1.3→80 Å / Num. obs: 58358 / % possible obs: 88 % / Redundancy: 31 % / Biso Wilson estimate: 19.2 Å2 / CC1/2: 1 / Rmerge(I) obs: 0.049 / Rpim(I) all: 0.008 / Rrim(I) all: 0.05 / Rsym value: 0.049 / Net I/σ(I): 42.7 |
| Reflection shell | Resolution: 1.3→1.33 Å / Redundancy: 4.2 % / Rmerge(I) obs: 0.36 / Mean I/σ(I) obs: 2.6 / Num. unique obs: 463 / CC1/2: 0.81 / Rpim(I) all: 0.205 / Rrim(I) all: 0.461 / Rsym value: 0.36 / % possible all: 14.4 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4EA6 Resolution: 1.3→80 Å / SU ML: 0.0786 / Cross valid method: FREE R-VALUE / σ(F): 1.37 / Phase error: 12.3452 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2 Details: Structure was thoroughly refined using REFMAC5 from CCP4 but then refined further using REFINE from PHENIX. Some improvements, such as addition of aa 241-244 at the cleavage site were made ...Details: Structure was thoroughly refined using REFMAC5 from CCP4 but then refined further using REFINE from PHENIX. Some improvements, such as addition of aa 241-244 at the cleavage site were made in REFINE. The restraints for the diglyceride substrate were made in the GRADE server. Waters were added by hand.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 21.44 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.3→80 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj