| Entry | Database: PDB / ID: 6uyb
|
|---|
| Title | Crystal structure of TEAD2 bound to Compound 1 |
|---|
 Components Components | Transcriptional enhancer factor TEF-4 |
|---|
 Keywords Keywords | Transcription/Transcription Inhibitor / EAD / YAP Binding Domain / Transcription Inhibitor / Palmitoylation inhibitor / Transcription-Transcription Inhibitor complex |
|---|
| Function / homology |  Function and homology information Function and homology information
TEAD-YAP complex / lateral mesoderm development / RUNX3 regulates YAP1-mediated transcription / notochord development / YAP1- and WWTR1 (TAZ)-stimulated gene expression / paraxial mesoderm development / hippo signaling / Formation of axial mesoderm / regulation of stem cell differentiation / embryonic heart tube morphogenesis ...TEAD-YAP complex / lateral mesoderm development / RUNX3 regulates YAP1-mediated transcription / notochord development / YAP1- and WWTR1 (TAZ)-stimulated gene expression / paraxial mesoderm development / hippo signaling / Formation of axial mesoderm / regulation of stem cell differentiation / embryonic heart tube morphogenesis / embryonic organ development / vasculogenesis / cellular response to retinoic acid / neural tube closure / transcription coactivator binding / disordered domain specific binding / sequence-specific double-stranded DNA binding / protein-containing complex assembly / transcription regulator complex / transcription by RNA polymerase II / DNA-binding transcription factor activity, RNA polymerase II-specific / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / intracellular membrane-bounded organelle / regulation of transcription by RNA polymerase II / regulation of DNA-templated transcription / positive regulation of DNA-templated transcription / chromatin / positive regulation of transcription by RNA polymerase II / nucleoplasm / nucleus / cytosolSimilarity search - Function Coagulation Factor XIII; Chain A, domain 1 - #80 / TEA/ATTS domain / Transcriptional enhancer factor, metazoa / TEA/ATTS domain superfamily / TEA/ATTS domain / TEA domain signature. / TEA domain profile. / TEA domain / YAP binding domain / : ...Coagulation Factor XIII; Chain A, domain 1 - #80 / TEA/ATTS domain / Transcriptional enhancer factor, metazoa / TEA/ATTS domain superfamily / TEA/ATTS domain / TEA domain signature. / TEA domain profile. / TEA domain / YAP binding domain / : / YAP binding domain / Coagulation Factor XIII; Chain A, domain 1 / Distorted Sandwich / Mainly BetaSimilarity search - Domain/homology |
|---|
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.543 Å MOLECULAR REPLACEMENT / Resolution: 1.543 Å |
|---|
 Authors Authors | Noland, C.L. / Holden, J.K. / Crawford, J.J. / Zbieg, J.R. / Cunningham, C.N. |
|---|
 Citation Citation |  Journal: Cell Rep / Year: 2020 Journal: Cell Rep / Year: 2020
Title: Small Molecule Dysregulation of TEAD Lipidation Induces a Dominant-Negative Inhibition of Hippo Pathway Signaling.
Authors: Holden, J.K. / Crawford, J.J. / Noland, C.L. / Schmidt, S. / Zbieg, J.R. / Lacap, J.A. / Zang, R. / Miller, G.M. / Zhang, Y. / Beroza, P. / Reja, R. / Lee, W. / Tom, J.Y.K. / Fong, R. / ...Authors: Holden, J.K. / Crawford, J.J. / Noland, C.L. / Schmidt, S. / Zbieg, J.R. / Lacap, J.A. / Zang, R. / Miller, G.M. / Zhang, Y. / Beroza, P. / Reja, R. / Lee, W. / Tom, J.Y.K. / Fong, R. / Steffek, M. / Clausen, S. / Hagenbeek, T.J. / Hu, T. / Zhou, Z. / Shen, H.C. / Cunningham, C.N. |
|---|
| History | | Deposition | Nov 12, 2019 | Deposition site: RCSB / Processing site: RCSB |
|---|
| Revision 1.0 | Jun 24, 2020 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Jul 8, 2020 | Group: Database references / Category: citation
Item: _citation.journal_volume / _citation.page_first ..._citation.journal_volume / _citation.page_first / _citation.page_last / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title |
|---|
| Revision 1.2 | Oct 11, 2023 | Group: Data collection / Database references / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / software
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _software.name |
|---|
|
|---|
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.543 Å
MOLECULAR REPLACEMENT / Resolution: 1.543 Å  Authors
Authors Citation
Citation Journal: Cell Rep / Year: 2020
Journal: Cell Rep / Year: 2020 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6uyb.cif.gz
6uyb.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6uyb.ent.gz
pdb6uyb.ent.gz PDB format
PDB format 6uyb.json.gz
6uyb.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/uy/6uyb
https://data.pdbj.org/pub/pdb/validation_reports/uy/6uyb ftp://data.pdbj.org/pub/pdb/validation_reports/uy/6uyb
ftp://data.pdbj.org/pub/pdb/validation_reports/uy/6uyb

 Links
Links Assembly
Assembly
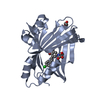

 Components
Components Homo sapiens (human) / Gene: TEAD2, TEF4 / Production host:
Homo sapiens (human) / Gene: TEAD2, TEF4 / Production host: 
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  ALS
ALS  / Beamline: 5.0.2 / Wavelength: 0.999 Å
/ Beamline: 5.0.2 / Wavelength: 0.999 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller













 PDBj
PDBj





