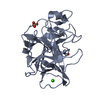
Entry Database : PDB / ID : 6th7Title Structure of porcine pancreatic elastase in complex with tutuilamide (Tutuilamide) x 2 Chymotrypsin-like elastase family member 1 Keywords / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / Biological species Sus scrofa (pig)Synthetic construct (others) Method / / / Resolution : 2.2 Å Authors Koehnke, J. / Sikandar, A. Funding support Organization Grant number Country German Research Foundation (DFG) KO4116_3_1
Journal : Acs Chem.Biol. / Year : 2020Title : Tutuilamides A-C: Vinyl-Chloride-Containing Cyclodepsipeptides from Marine Cyanobacteria with Potent Elastase Inhibitory Properties.Authors : Keller, L. / Canuto, K.M. / Liu, C. / Suzuki, B.M. / Almaliti, J. / Sikandar, A. / Naman, C.B. / Glukhov, E. / Luo, D. / Duggan, B.M. / Luesch, H. / Koehnke, J. / O'Donoghue, A.J. / Gerwick, W.H. History Deposition Nov 18, 2019 Deposition site / Processing site Revision 1.0 Mar 4, 2020 Provider / Type Revision 1.1 Apr 1, 2020 Group / Category / citation_authorItem _citation.journal_volume / _citation.page_first ... _citation.journal_volume / _citation.page_first / _citation.page_last / _citation_author.identifier_ORCID Revision 2.0 Dec 28, 2022 Group Advisory / Atomic model ... Advisory / Atomic model / Data collection / Database references / Derived calculations / Polymer sequence / Source and taxonomy / Structure summary Category atom_site / database_2 ... atom_site / database_2 / database_PDB_caveat / entity / entity_poly / entity_poly_seq / entity_src_nat / pdbx_entity_nonpoly / pdbx_entity_src_syn / pdbx_nonpoly_scheme / pdbx_poly_seq_scheme / pdbx_struct_assembly / pdbx_struct_assembly_gen / pdbx_struct_conn_angle / pdbx_validate_chiral / pdbx_validate_rmsd_bond / pdbx_validate_torsion / struct_asym / struct_conn / struct_conn_type / struct_mon_prot_cis / struct_ref / struct_ref_seq / struct_site / struct_site_gen Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _database_PDB_caveat.text / _entity_src_nat.common_name / _pdbx_struct_assembly.oligomeric_count / _pdbx_struct_assembly.oligomeric_details / _pdbx_struct_assembly_gen.asym_id_list / _pdbx_struct_conn_angle.ptnr2_auth_seq_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_validate_chiral.auth_asym_id / _pdbx_validate_chiral.auth_seq_id / _struct_conn.conn_type_id / _struct_conn.id / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_leaving_atom_flag / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_conn_type.id / _struct_site.details / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_seq_id / _struct_site_gen.auth_asym_id / _struct_site_gen.auth_seq_id / _struct_site_gen.label_asym_id / _struct_site_gen.label_seq_id Revision 3.0 Nov 15, 2023 Group / Data collection / Derived calculationsCategory atom_site / chem_comp_atom ... atom_site / chem_comp_atom / chem_comp_bond / struct_conn Item _atom_site.auth_atom_id / _atom_site.label_atom_id ... _atom_site.auth_atom_id / _atom_site.label_atom_id / _struct_conn.pdbx_leaving_atom_flag / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr2_label_atom_id Revision 3.1 Jan 31, 2024 Group / Category Revision 4.0 May 1, 2024 Group / Category / Item Revision 5.0 Jun 26, 2024 Group Atomic model / Data collection ... Atomic model / Data collection / Derived calculations / Non-polymer description / Polymer sequence / Structure summary Category atom_site / chem_comp ... atom_site / chem_comp / chem_comp_atom / chem_comp_bond / entity / entity_poly / entity_poly_seq / pdbx_poly_seq_scheme / pdbx_validate_rmsd_bond / struct_conn / struct_mon_prot_cis / struct_site / struct_site_gen Item _atom_site.Cartn_x / _atom_site.Cartn_y ... _atom_site.Cartn_x / _atom_site.Cartn_y / _atom_site.Cartn_z / _atom_site.auth_comp_id / _atom_site.group_PDB / _atom_site.label_comp_id / _chem_comp.formula / _chem_comp.formula_weight / _chem_comp.id / _chem_comp.mon_nstd_flag / _chem_comp.name / _entity.formula_weight / _entity_poly.pdbx_seq_one_letter_code / _entity_poly_seq.mon_id / _pdbx_poly_seq_scheme.mon_id / _pdbx_poly_seq_scheme.pdb_mon_id / _pdbx_validate_rmsd_bond.auth_comp_id_1 / _pdbx_validate_rmsd_bond.auth_comp_id_2 / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_label_comp_id / _struct_mon_prot_cis.auth_comp_id / _struct_mon_prot_cis.label_comp_id / _struct_site.pdbx_auth_comp_id / _struct_site_gen.auth_comp_id / _struct_site_gen.label_comp_id Revision 6.0 Jul 10, 2024 Group / Non-polymer description / Structure summaryCategory chem_comp / chem_comp_atom ... chem_comp / chem_comp_atom / chem_comp_bond / entity Item / _chem_comp.formula_weight / _entity.formula_weightRevision 7.0 Dec 24, 2025 Group Atomic model / Data collection ... Atomic model / Data collection / Derived calculations / Non-polymer description / Polymer sequence / Structure summary Category atom_site / chem_comp ... atom_site / chem_comp / chem_comp_atom / chem_comp_bond / entity_poly / entity_poly_seq / pdbx_entry_details / pdbx_modification_feature / pdbx_poly_seq_scheme / struct_conn / struct_site / struct_site_gen Item _atom_site.B_iso_or_equiv / _atom_site.Cartn_x ... _atom_site.B_iso_or_equiv / _atom_site.Cartn_x / _atom_site.Cartn_y / _atom_site.Cartn_z / _atom_site.auth_atom_id / _atom_site.auth_comp_id / _atom_site.label_atom_id / _atom_site.label_comp_id / _atom_site.type_symbol / _chem_comp.formula / _chem_comp.formula_weight / _chem_comp.id / _chem_comp.mon_nstd_flag / _chem_comp.name / _chem_comp.type / _entity_poly.pdbx_seq_one_letter_code / _entity_poly_seq.mon_id / _pdbx_entry_details.has_protein_modification / _pdbx_poly_seq_scheme.mon_id / _pdbx_poly_seq_scheme.pdb_mon_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_label_comp_id / _struct_site.pdbx_auth_comp_id / _struct_site_gen.auth_comp_id / _struct_site_gen.label_comp_id
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å
MOLECULAR REPLACEMENT / Resolution: 2.2 Å  Authors
Authors Germany, 1items
Germany, 1items  Citation
Citation Journal: Acs Chem.Biol. / Year: 2020
Journal: Acs Chem.Biol. / Year: 2020 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6th7.cif.gz
6th7.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6th7.ent.gz
pdb6th7.ent.gz PDB format
PDB format 6th7.json.gz
6th7.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/th/6th7
https://data.pdbj.org/pub/pdb/validation_reports/th/6th7 ftp://data.pdbj.org/pub/pdb/validation_reports/th/6th7
ftp://data.pdbj.org/pub/pdb/validation_reports/th/6th7
 Links
Links Assembly
Assembly


 Components
Components
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  ESRF
ESRF  / Beamline: MASSIF-3 / Wavelength: 0.9677 Å
/ Beamline: MASSIF-3 / Wavelength: 0.9677 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller











 PDBj
PDBj



