Entry Database : PDB / ID : 6t5zTitle Crystal structure of an AA10 LPMO from Photorhabdus luminescens Chitin-binding type-4 domain-containing protein Keywords / / / Function / homology / / / / / / Biological species Photorhabdus laumondii subsp. laumondii (bacteria)Method / / / Resolution : 1.6000030918 Å Authors Munzone, A. / El Kerdi, B. / Reglier, M. / Royant, A. / Simaan, A.J. / Decroos, C. Journal : Febs J. / Year : 2020Title : Characterization of a bacterial copper-dependent lytic polysaccharide monooxygenase with an unusual second coordination sphere.Authors : Munzone, A. / El Kerdi, B. / Fanuel, M. / Rogniaux, H. / Ropartz, D. / Reglier, M. / Royant, A. / Simaan, A.J. / Decroos, C. History Deposition Oct 17, 2019 Deposition site / Processing site Revision 1.0 Jan 15, 2020 Provider / Type Revision 1.1 Jan 22, 2020 Group Category / pdbx_struct_assembly_gen / struct_site_genItem _pdbx_struct_assembly.details / _pdbx_struct_assembly.method_details ... _pdbx_struct_assembly.details / _pdbx_struct_assembly.method_details / _pdbx_struct_assembly_gen.asym_id_list / _struct_site_gen.auth_comp_id / _struct_site_gen.auth_seq_id / _struct_site_gen.label_asym_id / _struct_site_gen.label_comp_id / _struct_site_gen.label_seq_id / _struct_site_gen.symmetry Revision 1.2 Aug 12, 2020 Group / Derived calculations / Category / pdbx_struct_conn_angle / struct_connItem _citation.journal_volume / _citation.page_first ... _citation.journal_volume / _citation.page_first / _citation.page_last / _pdbx_struct_conn_angle.ptnr1_auth_asym_id / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr1_symmetry / _pdbx_struct_conn_angle.ptnr2_auth_asym_id / _pdbx_struct_conn_angle.ptnr2_auth_seq_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr2_symmetry / _pdbx_struct_conn_angle.ptnr3_auth_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.ptnr3_symmetry / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_ptnr1_label_alt_id / _struct_conn.pdbx_ptnr2_label_alt_id / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr1_symmetry / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_conn.ptnr2_symmetry Revision 1.3 Jan 24, 2024 Group / Database references / Refinement descriptionCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model Item / _database_2.pdbx_database_accessionRevision 1.4 Nov 20, 2024 Group / Category / pdbx_modification_feature / Item
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Photorhabdus laumondii subsp. laumondii (bacteria)
Photorhabdus laumondii subsp. laumondii (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.6000030918 Å
MOLECULAR REPLACEMENT / Resolution: 1.6000030918 Å  Authors
Authors Citation
Citation Journal: Febs J. / Year: 2020
Journal: Febs J. / Year: 2020 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6t5z.cif.gz
6t5z.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6t5z.ent.gz
pdb6t5z.ent.gz PDB format
PDB format 6t5z.json.gz
6t5z.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/t5/6t5z
https://data.pdbj.org/pub/pdb/validation_reports/t5/6t5z ftp://data.pdbj.org/pub/pdb/validation_reports/t5/6t5z
ftp://data.pdbj.org/pub/pdb/validation_reports/t5/6t5z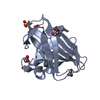
 Links
Links Assembly
Assembly



 Components
Components Photorhabdus laumondii subsp. laumondii (strain DSM 15139 / CIP 105565 / TT01) (bacteria)
Photorhabdus laumondii subsp. laumondii (strain DSM 15139 / CIP 105565 / TT01) (bacteria)
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  ESRF
ESRF  / Beamline: ID29 / Wavelength: 0.976 Å
/ Beamline: ID29 / Wavelength: 0.976 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller





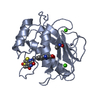







 PDBj
PDBj

