[English] 日本語
 Yorodumi
Yorodumi- PDB-6l76: Crystal structure of the Ni(II)(Chro)2-d(TTGGGCCGAA/TTCGGCCCAA) c... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6l76 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the Ni(II)(Chro)2-d(TTGGGCCGAA/TTCGGCCCAA) complex at 2.94 angstrom resolution | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA / Duplex DNA / GGGGCC motif / nucleotide flipping-out / Hoogsteen base pair | ||||||
| Function / homology | Chem-CPH / NICKEL (II) ION / DNA Function and homology information Function and homology information | ||||||
| Biological species | synthetic construct (others) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.94 Å MOLECULAR REPLACEMENT / Resolution: 2.94 Å | ||||||
 Authors Authors | Hou, M.H. / Jhan, C.R. / Satange, R.B. / Lin, S.M. | ||||||
 Citation Citation |  Journal: Nucleic Acids Res. / Year: 2021 Journal: Nucleic Acids Res. / Year: 2021Title: Targeting the ALS/FTD-associated A-DNA kink with anthracene-based metal complex causes DNA backbone straightening and groove contraction. Authors: Jhan, C.R. / Satange, R. / Wang, S.C. / Zeng, J.Y. / Horng, Y.C. / Jin, P. / Neidle, S. / Hou, M.H. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6l76.cif.gz 6l76.cif.gz | 77.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6l76.ent.gz pdb6l76.ent.gz | 57.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6l76.json.gz 6l76.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/l7/6l76 https://data.pdbj.org/pub/pdb/validation_reports/l7/6l76 ftp://data.pdbj.org/pub/pdb/validation_reports/l7/6l76 ftp://data.pdbj.org/pub/pdb/validation_reports/l7/6l76 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6l75C  7bpvC  1vaqS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 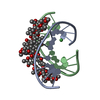
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-DNA chain , 2 types, 4 molecules ADBC
| #1: DNA chain | Mass: 3085.028 Da / Num. of mol.: 2 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) #2: DNA chain | Mass: 3004.980 Da / Num. of mol.: 2 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) |
|---|
-Sugars , 2 types, 8 molecules
| #3: Polysaccharide | 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H- ...2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate Source method: isolated from a genetically manipulated source #4: Polysaccharide | 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol- ...3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol Source method: isolated from a genetically manipulated source |
|---|
-Non-polymers , 3 types, 53 molecules 




| #5: Chemical | ChemComp-CPH / ( #6: Chemical | ChemComp-NI / #7: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | N |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 6.12 Å3/Da / Density % sol: 79.9 % |
|---|---|
| Crystal grow | Temperature: 277.15 K / Method: vapor diffusion, sitting drop / pH: 7 Details: 1mM TTGGGCCGAA, 1 mM TTCGGCCCAA, 3 mM Chromomycin A3, 6 mM NiSO4, 2.5 mM Spermine,40 mM Sodium cacodylate (pH=7), 4 % PEG 400 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSRRC NSRRC  / Beamline: BL15A1 / Wavelength: 0.99984 Å / Beamline: BL15A1 / Wavelength: 0.99984 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: RAYONIX MX300HE / Detector: CCD / Date: Mar 9, 2016 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.99984 Å / Relative weight: 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.94→30 Å / Num. obs: 12146 / % possible obs: 98.6 % / Redundancy: 9.3 % / Biso Wilson estimate: 74.54 Å2 / Rmerge(I) obs: 0.067 / Rpim(I) all: 0.023 / Rrim(I) all: 0.071 / Χ2: 0.909 / Net I/σ(I): 12.3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1VAQ Resolution: 2.94→25.511 Å / SU ML: 0.33 / Cross valid method: THROUGHOUT / σ(F): 1.34 / Phase error: 25.87
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 214.51 Å2 / Biso mean: 93.8655 Å2 / Biso min: 61.48 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.94→25.511 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: -26.1521 Å / Origin y: 6.1794 Å / Origin z: -14.636 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller









 PDBj
PDBj



