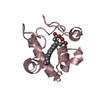
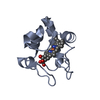
Entry Database : PDB / ID : 6k0lTitle The crystal structure of simian CD163 SRCR5 Scavenger receptor cysteine-rich type 1 protein M130 Keywords / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / Biological species Chlorocebus aethiops (grivet monkey)Method / / / Resolution : 1.58 Å Authors Ma, H. / Li, R. / Jiang, L. / Qiao, S. / Zhang, G. Funding support Organization Grant number Country National Natural Science Foundation of China 31490601 National Natural Science Foundation of China 31602036
Journal : Vet Res / Year : 2021Title : Structural comparison of CD163 SRCR5 from different species sheds some light on its involvement in porcine reproductive and respiratory syndrome virus-2 infection in vitro.Authors : Ma, H. / Li, R. / Jiang, L. / Qiao, S. / Chen, X.X. / Wang, A. / Zhang, G. History Deposition May 7, 2019 Deposition site / Processing site Revision 1.0 May 13, 2020 Provider / Type Revision 1.1 Nov 24, 2021 Group / Category / citation_author / database_2Item _citation.journal_abbrev / _citation.journal_id_CSD ... _citation.journal_abbrev / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.journal_volume / _citation.page_first / _citation.page_last / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year / _database_2.pdbx_DOI / _database_2.pdbx_database_accession Revision 1.2 Nov 22, 2023 Group / Refinement descriptionCategory / chem_comp_bond / pdbx_initial_refinement_modelRevision 1.3 Nov 20, 2024 Group / Category / pdbx_modification_feature
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Chlorocebus aethiops (grivet monkey)
Chlorocebus aethiops (grivet monkey) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.58 Å
MOLECULAR REPLACEMENT / Resolution: 1.58 Å  Authors
Authors China, 2items
China, 2items  Citation
Citation Journal: Vet Res / Year: 2021
Journal: Vet Res / Year: 2021 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6k0l.cif.gz
6k0l.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6k0l.ent.gz
pdb6k0l.ent.gz PDB format
PDB format 6k0l.json.gz
6k0l.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/k0/6k0l
https://data.pdbj.org/pub/pdb/validation_reports/k0/6k0l ftp://data.pdbj.org/pub/pdb/validation_reports/k0/6k0l
ftp://data.pdbj.org/pub/pdb/validation_reports/k0/6k0l

 Links
Links Assembly
Assembly


 Components
Components Chlorocebus aethiops (grivet monkey) / Gene: CD163, M130 / Production host:
Chlorocebus aethiops (grivet monkey) / Gene: CD163, M130 / Production host:  Komagataella pastoris (fungus) / References: UniProt: Q2VLG4
Komagataella pastoris (fungus) / References: UniProt: Q2VLG4 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SSRF
SSRF  / Beamline: BL18U1 / Wavelength: 0.979 Å
/ Beamline: BL18U1 / Wavelength: 0.979 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller











 PDBj
PDBj