[English] 日本語
 Yorodumi
Yorodumi- PDB-6g4z: Crystal structure of murine NF-kappaB inducing kinase (NIK) in co... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6g4z | ||||||
|---|---|---|---|---|---|---|---|
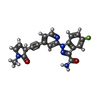
| Title | Crystal structure of murine NF-kappaB inducing kinase (NIK) in complex with compound 2f | ||||||
 Components Components | Mitogen-activated protein kinase kinase kinase 14 | ||||||
 Keywords Keywords | TRANSFERASE / PROTEIN SERINE/THREONINE KINASE / NF-KAPPAB / MAP3K14 | ||||||
| Function / homology |  Function and homology information Function and homology informationTNF receptor superfamily (TNFSF) members mediating non-canonical NF-kB pathway / TNFR2 non-canonical NF-kB pathway / Dectin-1 mediated noncanonical NF-kB signaling / NIK-->noncanonical NF-kB signaling / CD28 dependent PI3K/Akt signaling / mitogen-activated protein kinase kinase kinase / non-canonical NF-kappaB signal transduction / MAP kinase kinase kinase activity / cellular response to mechanical stimulus / fibrillar center ...TNF receptor superfamily (TNFSF) members mediating non-canonical NF-kB pathway / TNFR2 non-canonical NF-kB pathway / Dectin-1 mediated noncanonical NF-kB signaling / NIK-->noncanonical NF-kB signaling / CD28 dependent PI3K/Akt signaling / mitogen-activated protein kinase kinase kinase / non-canonical NF-kappaB signal transduction / MAP kinase kinase kinase activity / cellular response to mechanical stimulus / fibrillar center / defense response to virus / protein kinase activity / immune response / protein serine kinase activity / protein serine/threonine kinase activity / nucleoplasm / ATP binding / cytosol Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.84 Å MOLECULAR REPLACEMENT / Resolution: 2.84 Å | ||||||
 Authors Authors | Leonardo-Silvestre, H. / McEwan, P.A. / Hymowitz, S.G. | ||||||
 Citation Citation |  Journal: J. Med. Chem. / Year: 2018 Journal: J. Med. Chem. / Year: 2018Title: Scaffold-Hopping Approach To Discover Potent, Selective, and Efficacious Inhibitors of NF-kappa B Inducing Kinase. Authors: Blaquiere, N. / Castanedo, G.M. / Burch, J.D. / Berezhkovskiy, L.M. / Brightbill, H. / Brown, S. / Chan, C. / Chiang, P.C. / Crawford, J.J. / Dong, T. / Fan, P. / Feng, J. / Ghilardi, N. / ...Authors: Blaquiere, N. / Castanedo, G.M. / Burch, J.D. / Berezhkovskiy, L.M. / Brightbill, H. / Brown, S. / Chan, C. / Chiang, P.C. / Crawford, J.J. / Dong, T. / Fan, P. / Feng, J. / Ghilardi, N. / Godemann, R. / Gogol, E. / Grabbe, A. / Hole, A.J. / Hu, B. / Hymowitz, S.G. / Alaoui Ismaili, M.H. / Le, H. / Lee, P. / Lee, W. / Lin, X. / Liu, N. / McEwan, P.A. / McKenzie, B. / Silvestre, H.L. / Suto, E. / Sujatha-Bhaskar, S. / Wu, G. / Wu, L.C. / Zhang, Y. / Zhong, Z. / Staben, S.T. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6g4z.cif.gz 6g4z.cif.gz | 261 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6g4z.ent.gz pdb6g4z.ent.gz | 214.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6g4z.json.gz 6g4z.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/g4/6g4z https://data.pdbj.org/pub/pdb/validation_reports/g4/6g4z ftp://data.pdbj.org/pub/pdb/validation_reports/g4/6g4z ftp://data.pdbj.org/pub/pdb/validation_reports/g4/6g4z | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6g4yC  4g3eS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 38543.449 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: Q9WUL6, mitogen-activated protein kinase kinase kinase #2: Chemical | #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.17 Å3/Da / Density % sol: 61.14 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 6.2 Details: 0.3-0.9M ammonium sulphate, 0.05-0.1M sodium citrate, 0.7-1.0M Lithium sulphate |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I02 / Wavelength: 0.97949 Å / Beamline: I02 / Wavelength: 0.97949 Å |
| Detector | Type: DECTRIS PILATUS 6M-F / Detector: PIXEL / Date: Jun 12, 2013 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97949 Å / Relative weight: 1 |
| Reflection | Resolution: 2.84→36.21 Å / Num. obs: 21715 / % possible obs: 98.1 % / Redundancy: 6.1 % / CC1/2: 0.981 / Rmerge(I) obs: 0.104 / Net I/σ(I): 11.7 |
| Reflection shell | Resolution: 2.84→2.91 Å / Redundancy: 6.1 % / Rmerge(I) obs: 0.745 / Num. unique obs: 10567 / CC1/2: 0.84 / % possible all: 99.9 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4G3E Resolution: 2.84→36.21 Å / Cor.coef. Fo:Fc: 0.894 / Cor.coef. Fo:Fc free: 0.876 / SU B: 39.116 / SU ML: 0.342 / Cross valid method: THROUGHOUT / ESU R: 1.348 / ESU R Free: 0.4 / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 76.77 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.84→36.21 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj