| Entry | Database: PDB / ID: 6dnv
|
|---|
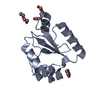
| Title | Crystal Structure of Neisseria meningitidis DsbD n-terminal domain in the reduced form |
|---|
 Components Components | Thiol:disulfide interchange protein DsbD |
|---|
 Keywords Keywords | OXIDOREDUCTASE / disulphide reductase / Dsb proteins |
|---|
| Function / homology |  Function and homology information Function and homology information
protein-disulfide reductase / protein-disulfide reductase [NAD(P)H] activity / cytochrome complex assembly / cell redox homeostasis / electron transfer activity / metal ion binding / plasma membraneSimilarity search - Function Thiol:disulfide interchange protein DsbD, N-terminal domain / Thiol:disulphide interchange protein DsbD / Thiol:disulfide interchange protein DsbD, N-terminal domain superfamily / DsbD gamma / Thiol:disulfide interchange protein DsbD, N-terminal domain / Thiol:disulfide interchange protein DsbD, N-terminal / Cytochrome C biogenesis protein, transmembrane domain / Cytochrome C biogenesis protein transmembrane domain / Thioredoxin-like domain / Thioredoxin-like fold ...Thiol:disulfide interchange protein DsbD, N-terminal domain / Thiol:disulphide interchange protein DsbD / Thiol:disulfide interchange protein DsbD, N-terminal domain superfamily / DsbD gamma / Thiol:disulfide interchange protein DsbD, N-terminal domain / Thiol:disulfide interchange protein DsbD, N-terminal / Cytochrome C biogenesis protein, transmembrane domain / Cytochrome C biogenesis protein transmembrane domain / Thioredoxin-like domain / Thioredoxin-like fold / Thioredoxin domain profile. / Thioredoxin domain / Thioredoxin-like superfamily / Immunoglobulin-like / Sandwich / Mainly BetaSimilarity search - Domain/homology |
|---|
| Biological species |  Neisseria meningitidis (bacteria) Neisseria meningitidis (bacteria) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.603 Å molecular replacement / Resolution: 1.603 Å |
|---|
 Authors Authors | Smith, R.P. / Heras, B. / Paxman, J.J. |
|---|
 Citation Citation |  Journal: J. Biol. Chem. / Year: 2018Title Journal: J. Biol. Chem. / Year: 2018Title: Structural and biochemical insights into the disulfide reductase mechanism of DsbD, an essential enzyme for neisserial pathogens. Authors: Smith, R.P. / Mohanty, B. / Mowlaboccus, S. / Paxman, J.J. / Williams, M.L. / Headey, S.J. / Wang, G. / Subedi, P. / Doak, B.C. / Kahler, C.M. / Scanlon, M.J. / Heras, B. |
|---|
| History | | Deposition | Jun 8, 2018 | Deposition site: RCSB / Processing site: RCSB |
|---|
| Revision 1.0 | Sep 12, 2018 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Sep 19, 2018 | Group: Data collection / Database references / Category: citation / citation_author
Item: _citation.country / _citation.journal_abbrev ..._citation.country / _citation.journal_abbrev / _citation.journal_id_ASTM / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year / _citation_author.identifier_ORCID |
|---|
| Revision 1.2 | Nov 7, 2018 | Group: Data collection / Database references / Category: citation
Item: _citation.journal_volume / _citation.page_first / _citation.page_last |
|---|
| Revision 1.3 | Mar 13, 2024 | Group: Data collection / Database references / Category: chem_comp_atom / chem_comp_bond / database_2
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Neisseria meningitidis (bacteria)
Neisseria meningitidis (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT /
MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.603 Å
molecular replacement / Resolution: 1.603 Å  Authors
Authors Citation
Citation Journal: J. Biol. Chem. / Year: 2018
Journal: J. Biol. Chem. / Year: 2018 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6dnv.cif.gz
6dnv.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6dnv.ent.gz
pdb6dnv.ent.gz PDB format
PDB format 6dnv.json.gz
6dnv.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/dn/6dnv
https://data.pdbj.org/pub/pdb/validation_reports/dn/6dnv ftp://data.pdbj.org/pub/pdb/validation_reports/dn/6dnv
ftp://data.pdbj.org/pub/pdb/validation_reports/dn/6dnv

 Links
Links Assembly
Assembly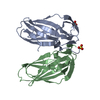


 Components
Components Neisseria meningitidis (bacteria) / Strain: alpha14 / Gene: dsbD, NMO_1340 / Plasmid: pMCSG7
Neisseria meningitidis (bacteria) / Strain: alpha14 / Gene: dsbD, NMO_1340 / Plasmid: pMCSG7
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  Australian Synchrotron
Australian Synchrotron  / Beamline: MX2 / Wavelength: 0.9537 Å
/ Beamline: MX2 / Wavelength: 0.9537 Å molecular replacement
molecular replacement Processing
Processing MOLECULAR REPLACEMENT / Resolution: 1.603→41.726 Å / SU ML: 0.19 / Cross valid method: THROUGHOUT / σ(F): 1.34 / Phase error: 19.26
MOLECULAR REPLACEMENT / Resolution: 1.603→41.726 Å / SU ML: 0.19 / Cross valid method: THROUGHOUT / σ(F): 1.34 / Phase error: 19.26  Movie
Movie Controller
Controller












 PDBj
PDBj

