+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6cnp | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of MBD2 complex with methylated CpG island | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | TRANSCRIPTION/DNA / Structural Genomics Consortium / SGC / TRANSCRIPTION-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsatellite DNA binding / ventricular cardiac muscle tissue development / NuRD complex / siRNA binding / maternal behavior / C2H2 zinc finger domain binding / methyl-CpG binding / DNA methylation-dependent constitutive heterochromatin formation / embryonic organ development / response to mechanical stimulus ...satellite DNA binding / ventricular cardiac muscle tissue development / NuRD complex / siRNA binding / maternal behavior / C2H2 zinc finger domain binding / methyl-CpG binding / DNA methylation-dependent constitutive heterochromatin formation / embryonic organ development / response to mechanical stimulus / positive regulation of Wnt signaling pathway / heterochromatin / RNA Polymerase I Promoter Opening / response to nutrient levels / NoRC negatively regulates rRNA expression / Wnt signaling pathway / response to estradiol / regulation of cell population proliferation / protein-containing complex assembly / molecular adaptor activity / chromatin remodeling / protein domain specific binding / negative regulation of DNA-templated transcription / mRNA binding / chromatin binding / chromatin / positive regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II / protein-containing complex / nucleoplasm / identical protein binding / nucleus / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human)synthetic construct (others) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.1 Å MOLECULAR REPLACEMENT / Resolution: 2.1 Å | |||||||||
 Authors Authors | Xu, C. / Min, J. / Structural Genomics Consortium (SGC) | |||||||||
 Citation Citation |  Journal: J. Biol. Chem. / Year: 2018 Journal: J. Biol. Chem. / Year: 2018Title: Structural basis for the ability of MBD domains to bind methyl-CG and TG sites in DNA. Authors: Liu, K. / Xu, C. / Lei, M. / Yang, A. / Loppnau, P. / Hughes, T.R. / Min, J. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6cnp.cif.gz 6cnp.cif.gz | 120.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6cnp.ent.gz pdb6cnp.ent.gz | 88.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6cnp.json.gz 6cnp.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  6cnp_validation.pdf.gz 6cnp_validation.pdf.gz | 452.1 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  6cnp_full_validation.pdf.gz 6cnp_full_validation.pdf.gz | 454.1 KB | Display | |
| Data in XML |  6cnp_validation.xml.gz 6cnp_validation.xml.gz | 8.9 KB | Display | |
| Data in CIF |  6cnp_validation.cif.gz 6cnp_validation.cif.gz | 11.8 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cn/6cnp https://data.pdbj.org/pub/pdb/validation_reports/cn/6cnp ftp://data.pdbj.org/pub/pdb/validation_reports/cn/6cnp ftp://data.pdbj.org/pub/pdb/validation_reports/cn/6cnp | HTTPS FTP |
-Related structure data
| Related structure data |  6c1aC  6c1tC  6c1uC  6c1vC  6cnqC  2ky8S  3qmgS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 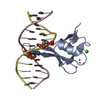
| ||||||||
| 2 | 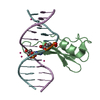
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 10934.497 Da / Num. of mol.: 2 / Fragment: residues 143-220 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: MBD2 / Plasmid: pET28-mhl / Production host: Homo sapiens (human) / Gene: MBD2 / Plasmid: pET28-mhl / Production host:  #2: DNA chain | Mass: 3678.407 Da / Num. of mol.: 4 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) #3: Chemical | ChemComp-CA / | #4: Chemical | ChemComp-UNX / #5: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.2 Å3/Da / Density % sol: 60.7 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, sitting drop / Details: 8% PEG550MME, 8% PEG20000, 0.2M calcium acetate |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 23-ID-D / Wavelength: 0.97947 Å / Beamline: 23-ID-D / Wavelength: 0.97947 Å | ||||||||||||||||||||||||
| Detector | Type: MARMOSAIC 300 mm CCD / Detector: CCD / Date: Jun 18, 2011 | ||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.97947 Å / Relative weight: 1 | ||||||||||||||||||||||||
| Reflection | Resolution: 2.1→39.46 Å / Num. obs: 20704 / % possible obs: 97.8 % / Redundancy: 3.9 % / Biso Wilson estimate: 50.39 Å2 / CC1/2: 0.998 / Rmerge(I) obs: 0.044 / Rpim(I) all: 0.026 / Rrim(I) all: 0.051 / Net I/σ(I): 15.3 / Num. measured all: 80533 / Scaling rejects: 0 | ||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: pdb entries 2KY8, 3QMG Resolution: 2.1→39.457 Å / SU ML: 0.22 / Cross valid method: FREE R-VALUE / σ(F): 1.92 / Phase error: 29.66 Details: Prior to molecular replacement, protein coordinates of the NMR model ensemble of pdb entry 2KY8 were averaged using a program written by Aiping Dong. refmac was used during intermediate ...Details: Prior to molecular replacement, protein coordinates of the NMR model ensemble of pdb entry 2KY8 were averaged using a program written by Aiping Dong. refmac was used during intermediate refinement steps. coot was used for interactive model building. Model geometry was assessed with molprobity.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 120.94 Å2 / Biso mean: 61.2521 Å2 / Biso min: 33.42 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.1→39.457 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 15
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj











































