| Entry | Database: PDB / ID: 5y41
|
|---|
| Title | Crystal Structure of LIGAND-BOUND NURR1-LBD |
|---|
 Components Components | Nuclear receptor subfamily 4 group A member 2 |
|---|
 Keywords Keywords | TRANSCRIPTION / Nurr1 / LBD / Prostaglandin / MICHAEL ADDITION |
|---|
| Function / homology |  Function and homology information Function and homology information
general adaptation syndrome / habenula development / cellular response to corticotropin-releasing hormone stimulus / central nervous system projection neuron axonogenesis / nuclear glucocorticoid receptor binding / regulation of dopamine metabolic process / dopaminergic neuron differentiation / midbrain dopaminergic neuron differentiation / neuron maturation / regulation of respiratory gaseous exchange ...general adaptation syndrome / habenula development / cellular response to corticotropin-releasing hormone stimulus / central nervous system projection neuron axonogenesis / nuclear glucocorticoid receptor binding / regulation of dopamine metabolic process / dopaminergic neuron differentiation / midbrain dopaminergic neuron differentiation / neuron maturation / regulation of respiratory gaseous exchange / central nervous system neuron differentiation / dopamine biosynthetic process / negative regulation of apoptotic signaling pathway / canonical Wnt signaling pathway / nuclear retinoid X receptor binding / fat cell differentiation / response to amphetamine / adult locomotory behavior / post-embryonic development / SUMOylation of intracellular receptors / beta-catenin binding / Nuclear Receptor transcription pathway / nuclear receptor activity / neuron migration / sequence-specific double-stranded DNA binding / DNA-binding transcription activator activity, RNA polymerase II-specific / cellular response to oxidative stress / neuron apoptotic process / transcription regulator complex / negative regulation of neuron apoptotic process / transcription by RNA polymerase II / DNA-binding transcription factor activity, RNA polymerase II-specific / response to hypoxia / nuclear speck / RNA polymerase II cis-regulatory region sequence-specific DNA binding / protein heterodimerization activity / regulation of transcription by RNA polymerase II / DNA-templated transcription / chromatin / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / protein-containing complex / DNA binding / zinc ion binding / nucleoplasm / nucleus / cytoplasmSimilarity search - Function Orphan nuclear receptor, NURR type / Orphan nuclear receptor / Retinoid X Receptor / Retinoid X Receptor / Nuclear hormone receptor / Nuclear hormones receptors DNA-binding region signature. / Zinc finger, nuclear hormone receptor-type / Double treble clef zinc finger, C4 type / Nuclear hormone receptors DNA-binding domain profile. / c4 zinc finger in nuclear hormone receptors ...Orphan nuclear receptor, NURR type / Orphan nuclear receptor / Retinoid X Receptor / Retinoid X Receptor / Nuclear hormone receptor / Nuclear hormones receptors DNA-binding region signature. / Zinc finger, nuclear hormone receptor-type / Double treble clef zinc finger, C4 type / Nuclear hormone receptors DNA-binding domain profile. / c4 zinc finger in nuclear hormone receptors / Nuclear hormone receptor, ligand-binding domain / Nuclear hormone receptor-like domain superfamily / Ligand-binding domain of nuclear hormone receptor / Nuclear receptor (NR) ligand-binding (LBD) domain profile. / Ligand binding domain of hormone receptors / Zinc finger, NHR/GATA-type / Orthogonal Bundle / Mainly AlphaSimilarity search - Domain/homology |
|---|
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.05 Å MOLECULAR REPLACEMENT / Resolution: 2.05 Å |
|---|
 Authors Authors | Sreekanth, R. / Lescar, J. / Yoon, H.S. |
|---|
 Citation Citation |  Journal: Nat.Chem.Biol. / Year: 2020 Journal: Nat.Chem.Biol. / Year: 2020
Title: PGE1 and PGA1 bind to Nurr1 and activate its transcriptional function.
Authors: Rajan, S. / Jang, Y. / Kim, C.H. / Kim, W. / Toh, H.T. / Jeon, J. / Song, B. / Serra, A. / Lescar, J. / Yoo, J.Y. / Beldar, S. / Ye, H. / Kang, C. / Liu, X.W. / Feitosa, M. / Kim, Y. / ...Authors: Rajan, S. / Jang, Y. / Kim, C.H. / Kim, W. / Toh, H.T. / Jeon, J. / Song, B. / Serra, A. / Lescar, J. / Yoo, J.Y. / Beldar, S. / Ye, H. / Kang, C. / Liu, X.W. / Feitosa, M. / Kim, Y. / Hwang, D. / Goh, G. / Lim, K.L. / Park, H.M. / Lee, C.H. / Oh, S.F. / Petsko, G.A. / Yoon, H.S. / Kim, K.S. |
|---|
| History | | Deposition | Jul 31, 2017 | Deposition site: PDBJ / Processing site: PDBJ |
|---|
| Revision 1.0 | Dec 26, 2018 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Jun 10, 2020 | Group: Database references / Category: citation / citation_author
Item: _citation.country / _citation.journal_abbrev ..._citation.country / _citation.journal_abbrev / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year |
|---|
| Revision 1.2 | Nov 22, 2023 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_conn / struct_ncs_dom_lim
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_conn.conn_type_id / _struct_conn.id / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_leaving_atom_flag / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr1_symmetry / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_conn.ptnr2_symmetry / _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id / _struct_ncs_dom_lim.beg_label_comp_id / _struct_ncs_dom_lim.beg_label_seq_id / _struct_ncs_dom_lim.end_auth_comp_id / _struct_ncs_dom_lim.end_label_asym_id / _struct_ncs_dom_lim.end_label_comp_id / _struct_ncs_dom_lim.end_label_seq_id |
|---|
| Revision 1.3 | Nov 6, 2024 | Group: Structure summary / Category: pdbx_entry_details / pdbx_modification_feature |
|---|
|
|---|
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.05 Å
MOLECULAR REPLACEMENT / Resolution: 2.05 Å  Authors
Authors Citation
Citation Journal: Nat.Chem.Biol. / Year: 2020
Journal: Nat.Chem.Biol. / Year: 2020 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5y41.cif.gz
5y41.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5y41.ent.gz
pdb5y41.ent.gz PDB format
PDB format 5y41.json.gz
5y41.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/y4/5y41
https://data.pdbj.org/pub/pdb/validation_reports/y4/5y41 ftp://data.pdbj.org/pub/pdb/validation_reports/y4/5y41
ftp://data.pdbj.org/pub/pdb/validation_reports/y4/5y41
 Links
Links Assembly
Assembly
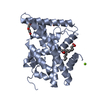

 Components
Components Homo sapiens (human) / Gene: NR4A2, NOT, NURR1, TINUR / Plasmid: PET-SUMO / Production host:
Homo sapiens (human) / Gene: NR4A2, NOT, NURR1, TINUR / Plasmid: PET-SUMO / Production host: 
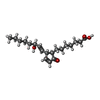










 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SLS
SLS  / Beamline: X06DA / Wavelength: 1.00003 Å
/ Beamline: X06DA / Wavelength: 1.00003 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj



