| Entry | Database: PDB / ID: 5wtl
|
|---|
| Title | Crystal structure of the periplasmic portion of outer membrane protein A (OmpA) from Capnocytophaga gingivalis |
|---|
 Components Components | OmpA family protein |
|---|
 Keywords Keywords | MEMBRANE PROTEIN / prokaryote / TT3R / TSP3 / Calcium-binding motif |
|---|
| Function / homology |  Function and homology information Function and homology information
porin activity / pore complex / monoatomic ion transport / cell outer membrane / cell adhesion / calcium ion bindingSimilarity search - Function Outer membrane protein beta-barrel domain / Outer membrane protein beta-barrel domain / Thrombospondin, type 3-like repeat / Thrombospondin type 3 repeat / TSP type-3 repeat / Outer membrane protein, bacterial / OmpA-like domain profile. / OmpA-like domain superfamily / OmpA family / OmpA-like domain ...Outer membrane protein beta-barrel domain / Outer membrane protein beta-barrel domain / Thrombospondin, type 3-like repeat / Thrombospondin type 3 repeat / TSP type-3 repeat / Outer membrane protein, bacterial / OmpA-like domain profile. / OmpA-like domain superfamily / OmpA family / OmpA-like domain / Outer membrane protein/outer membrane enzyme PagP, beta-barrel / EF-Hand 1, calcium-binding site / EF-hand calcium-binding domain.Similarity search - Domain/homology |
|---|
| Biological species |  Capnocytophaga gingivalis (bacteria) Capnocytophaga gingivalis (bacteria) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.298 Å MOLECULAR REPLACEMENT / Resolution: 2.298 Å |
|---|
 Authors Authors | Dai, S. / Tan, K. / Ye, S. / Zhang, R. |
|---|
 Citation Citation |  Journal: Cell Calcium / Year: 2017 Journal: Cell Calcium / Year: 2017
Title: Structure of thrombospondin type 3 repeats in bacterial outer membrane protein A reveals its intra-repeat disulfide bond-dependent calcium-binding capability.
Authors: Dai, S. / Sun, C. / Tan, K. / Ye, S. / Zhang, R. |
|---|
| History | | Deposition | Dec 13, 2016 | Deposition site: PDBJ / Processing site: PDBJ |
|---|
| Revision 1.0 | Dec 13, 2017 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Sep 12, 2018 | Group: Data collection / Database references / Category: citation / citation_author
Item: _citation.journal_abbrev / _citation.journal_id_CSD ..._citation.journal_abbrev / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.journal_volume / _citation.page_first / _citation.page_last / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year |
|---|
| Revision 1.2 | Nov 8, 2023 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / pdbx_struct_conn_angle / struct_conn
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_asym_id / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr3_auth_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id |
|---|
| Revision 1.3 | Nov 13, 2024 | Group: Database references / Structure summary
Category: citation / pdbx_entry_details / pdbx_modification_feature
Item: _citation.country |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Capnocytophaga gingivalis (bacteria)
Capnocytophaga gingivalis (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.298 Å
MOLECULAR REPLACEMENT / Resolution: 2.298 Å  Authors
Authors Citation
Citation Journal: Cell Calcium / Year: 2017
Journal: Cell Calcium / Year: 2017 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5wtl.cif.gz
5wtl.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5wtl.ent.gz
pdb5wtl.ent.gz PDB format
PDB format 5wtl.json.gz
5wtl.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/wt/5wtl
https://data.pdbj.org/pub/pdb/validation_reports/wt/5wtl ftp://data.pdbj.org/pub/pdb/validation_reports/wt/5wtl
ftp://data.pdbj.org/pub/pdb/validation_reports/wt/5wtl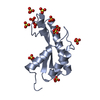

 Links
Links Assembly
Assembly




 Components
Components Capnocytophaga gingivalis (bacteria) / Gene: CAPGI0001_0903 / Production host:
Capnocytophaga gingivalis (bacteria) / Gene: CAPGI0001_0903 / Production host: 
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SSRF
SSRF  / Beamline: BL17U / Wavelength: 0.97931 Å
/ Beamline: BL17U / Wavelength: 0.97931 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj







