Entry Database : PDB / ID : 5t7kTitle X-ray crystal structure of AA13 LPMO AoAA13 Keywords / / Function / homology / / / / / Biological species Aspergillus oryzae RIB40 (mold)Method / / / Resolution : 1.3 Å Authors Frandsen, K.E.H. / Poulsen, J.-C.N. / Tovborg, M. / Johansen, K.S. / Lo Leggio, L. Funding support Organization Grant number Country The Danish Council for Strategic Research 12-134923
Journal : Acta Crystallogr D Struct Biol / Year : 2017Title : Learning from oligosaccharide soaks of crystals of an AA13 lytic polysaccharide monooxygenase: crystal packing, ligand binding and active-site disorder.Authors : Frandsen, K.E. / Poulsen, J.C. / Tovborg, M. / Johansen, K.S. / Lo Leggio, L. History Deposition Sep 5, 2016 Deposition site / Processing site Revision 1.0 Jan 18, 2017 Provider / Type Revision 1.1 Jan 17, 2018 Group / Category / Item Revision 1.2 Jul 29, 2020 Group / Derived calculations / Structure summaryCategory chem_comp / entity ... chem_comp / entity / pdbx_chem_comp_identifier / pdbx_entity_nonpoly / pdbx_struct_conn_angle / struct_conn / struct_conn_type / struct_site / struct_site_gen Item _chem_comp.name / _chem_comp.type ... _chem_comp.name / _chem_comp.type / _entity.pdbx_description / _pdbx_entity_nonpoly.name / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_alt_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_alt_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.conn_type_id / _struct_conn.id / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_leaving_atom_flag / _struct_conn.pdbx_ptnr1_label_alt_id / _struct_conn.pdbx_ptnr2_label_alt_id / _struct_conn.pdbx_role / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_conn.ptnr2_symmetry / _struct_conn_type.id Description / Provider / Type Revision 1.3 Jan 17, 2024 Group Data collection / Database references ... Data collection / Database references / Refinement description / Structure summary Category chem_comp / chem_comp_atom ... chem_comp / chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model Item / _database_2.pdbx_DOI / _database_2.pdbx_database_accession
Show all Show less
 Open data
Open data Basic information
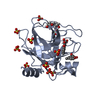
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.3 Å
MOLECULAR REPLACEMENT / Resolution: 1.3 Å  Authors
Authors Denmark, 1items
Denmark, 1items  Citation
Citation Journal: Acta Crystallogr D Struct Biol / Year: 2017
Journal: Acta Crystallogr D Struct Biol / Year: 2017 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5t7k.cif.gz
5t7k.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5t7k.ent.gz
pdb5t7k.ent.gz PDB format
PDB format 5t7k.json.gz
5t7k.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads 5t7k_validation.pdf.gz
5t7k_validation.pdf.gz wwPDB validaton report
wwPDB validaton report 5t7k_full_validation.pdf.gz
5t7k_full_validation.pdf.gz 5t7k_validation.xml.gz
5t7k_validation.xml.gz 5t7k_validation.cif.gz
5t7k_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/t7/5t7k
https://data.pdbj.org/pub/pdb/validation_reports/t7/5t7k ftp://data.pdbj.org/pub/pdb/validation_reports/t7/5t7k
ftp://data.pdbj.org/pub/pdb/validation_reports/t7/5t7k



 Links
Links Assembly
Assembly
 Components
Components

 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  MAX II
MAX II  / Beamline: I911-3 / Wavelength: 1 Å
/ Beamline: I911-3 / Wavelength: 1 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj



