| 登録情報 | データベース: PDB / ID: 1t31
|
|---|
| タイトル | A Dual Inhibitor of the Leukocyte Proteases Cathepsin G and Chymase with Therapeutic Efficacy in Animals Models of Inflammation |
|---|
 要素 要素 | Chymase |
|---|
 キーワード キーワード | HYDROLASE / HUMAN CHYMASE / SERINE PROTEINASE |
|---|
| 機能・相同性 |  機能・相同性情報 機能・相同性情報
chymase / basement membrane disassembly / cytokine precursor processing / peptide metabolic process / Activation of Matrix Metalloproteinases / midbrain development / extracellular matrix disassembly / Metabolism of Angiotensinogen to Angiotensins / angiotensin maturation / peptide binding ...chymase / basement membrane disassembly / cytokine precursor processing / peptide metabolic process / Activation of Matrix Metalloproteinases / midbrain development / extracellular matrix disassembly / Metabolism of Angiotensinogen to Angiotensins / angiotensin maturation / peptide binding / serine-type peptidase activity / secretory granule / protein maturation / protein catabolic process / cellular response to glucose stimulus / Signaling by SCF-KIT / : / cytoplasmic ribonucleoprotein granule / positive regulation of angiogenesis / regulation of inflammatory response / endopeptidase activity / serine-type endopeptidase activity / extracellular space / extracellular region / cytosol類似検索 - 分子機能 Serine proteases, trypsin family, histidine active site / Serine proteases, trypsin family, serine active site / Serine proteases, trypsin family, histidine active site. / Peptidase S1A, chymotrypsin family / Serine proteases, trypsin family, serine active site. / Serine proteases, trypsin domain profile. / Trypsin-like serine protease / Serine proteases, trypsin domain / Trypsin / Trypsin-like serine proteases ...Serine proteases, trypsin family, histidine active site / Serine proteases, trypsin family, serine active site / Serine proteases, trypsin family, histidine active site. / Peptidase S1A, chymotrypsin family / Serine proteases, trypsin family, serine active site. / Serine proteases, trypsin domain profile. / Trypsin-like serine protease / Serine proteases, trypsin domain / Trypsin / Trypsin-like serine proteases / Thrombin, subunit H / Peptidase S1, PA clan, chymotrypsin-like fold / Peptidase S1, PA clan / Beta Barrel / Mainly Beta類似検索 - ドメイン・相同性 |
|---|
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) |
|---|
| 手法 |  X線回折 / X線回折 /  シンクロトロン / シンクロトロン /  分子置換 / 解像度: 1.9 Å 分子置換 / 解像度: 1.9 Å |
|---|
 データ登録者 データ登録者 | de Garavilla, L. / Greco, M.N. / Giardino, E.C. / Wells, G.I. / Haertlein, B.J. / Kauffman, J.A. / Corcoran, T.W. / Derian, C.K. / Eckardt, A.J. / Abraham, W.M. ...de Garavilla, L. / Greco, M.N. / Giardino, E.C. / Wells, G.I. / Haertlein, B.J. / Kauffman, J.A. / Corcoran, T.W. / Derian, C.K. / Eckardt, A.J. / Abraham, W.M. / Sukumar, N. / Chen, Z. / Pineda, A.O. / Mathews, F.S. / Di Cera, E. / Andrade-Gordon, P. / Damiano, B.P. / Maryanoff, B.E. / Pereira, P.J.B. / Wang, Z.M. / Rubin, H. / Huber, R. / Bode, W. / Schechter, N.M. / Strobl, S. |
|---|
 引用 引用 |  ジャーナル: J.Biol.Chem. / 年: 2005 ジャーナル: J.Biol.Chem. / 年: 2005
タイトル: A novel, potent dual inhibitor of the leukocyte proteases cathepsin G and chymase: molecular mechanisms and anti-inflammatory activity in vivo.
著者: de Garavilla, L. / Greco, M.N. / Sukumar, N. / Chen, Z.W. / Pineda, A.O. / Mathews, F.S. / Di Cera, E. / Giardino, E.C. / Wells, G.I. / Haertlein, B.J. / Kauffman, J.A. / Corcoran, T.W. / ...著者: de Garavilla, L. / Greco, M.N. / Sukumar, N. / Chen, Z.W. / Pineda, A.O. / Mathews, F.S. / Di Cera, E. / Giardino, E.C. / Wells, G.I. / Haertlein, B.J. / Kauffman, J.A. / Corcoran, T.W. / Derian, C.K. / Eckardt, A.J. / Damiano, B.P. / Andrade-Gordon, P. / Maryanoff, B.E. |
|---|
| 履歴 | | 登録 | 2004年4月23日 | 登録サイト: RCSB / 処理サイト: RCSB |
|---|
| 改定 1.0 | 2005年3月1日 | Provider: repository / タイプ: Initial release |
|---|
| 改定 1.1 | 2008年4月30日 | Group: Version format compliance |
|---|
| 改定 1.2 | 2011年7月13日 | Group: Derived calculations / Version format compliance |
|---|
| 改定 2.0 | 2020年7月29日 | Group: Advisory / Atomic model ...Advisory / Atomic model / Data collection / Derived calculations / Structure summary
カテゴリ: atom_site / chem_comp ...atom_site / chem_comp / entity / pdbx_branch_scheme / pdbx_chem_comp_identifier / pdbx_entity_branch / pdbx_entity_branch_descriptor / pdbx_entity_branch_link / pdbx_entity_branch_list / pdbx_entity_nonpoly / pdbx_nonpoly_scheme / pdbx_struct_assembly_gen / pdbx_struct_conn_angle / pdbx_struct_special_symmetry / pdbx_validate_close_contact / struct_asym / struct_conn / struct_site / struct_site_gen
Item: _atom_site.auth_asym_id / _atom_site.auth_seq_id ..._atom_site.auth_asym_id / _atom_site.auth_seq_id / _atom_site.label_asym_id / _atom_site.label_entity_id / _chem_comp.name / _chem_comp.type / _pdbx_entity_nonpoly.entity_id / _pdbx_entity_nonpoly.name / _pdbx_struct_assembly_gen.asym_id_list / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr1_symmetry / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.ptnr3_symmetry / _pdbx_struct_conn_angle.value / _pdbx_struct_special_symmetry.label_asym_id / _pdbx_validate_close_contact.auth_asym_id_2 / _pdbx_validate_close_contact.auth_seq_id_2 / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_leaving_atom_flag / _struct_conn.pdbx_role / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr1_symmetry / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_conn.ptnr2_symmetry
解説: Carbohydrate remediation / Provider: repository / タイプ: Remediation |
|---|
| 改定 2.1 | 2021年10月27日 | Group: Database references / Structure summary / カテゴリ: chem_comp / database_2 / struct_ref_seq_dif
Item: _chem_comp.pdbx_synonyms / _database_2.pdbx_DOI ..._chem_comp.pdbx_synonyms / _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_ref_seq_dif.details |
|---|
| 改定 2.2 | 2023年8月23日 | Group: Data collection / Refinement description
カテゴリ: chem_comp_atom / chem_comp_bond / pdbx_initial_refinement_model |
|---|
| 改定 2.3 | 2024年10月30日 | Group: Structure summary
カテゴリ: pdbx_entry_details / pdbx_modification_feature |
|---|
|
|---|
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト)
Homo sapiens (ヒト) X線回折 /
X線回折 /  シンクロトロン /
シンクロトロン /  分子置換 / 解像度: 1.9 Å
分子置換 / 解像度: 1.9 Å  データ登録者
データ登録者 引用
引用 ジャーナル: J.Biol.Chem. / 年: 2005
ジャーナル: J.Biol.Chem. / 年: 2005 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 1t31.cif.gz
1t31.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb1t31.ent.gz
pdb1t31.ent.gz PDB形式
PDB形式 1t31.json.gz
1t31.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード https://data.pdbj.org/pub/pdb/validation_reports/t3/1t31
https://data.pdbj.org/pub/pdb/validation_reports/t3/1t31 ftp://data.pdbj.org/pub/pdb/validation_reports/t3/1t31
ftp://data.pdbj.org/pub/pdb/validation_reports/t3/1t31 リンク
リンク 集合体
集合体

 要素
要素 Homo sapiens (ヒト) / 遺伝子: CMA1, CYM, CYH
Homo sapiens (ヒト) / 遺伝子: CMA1, CYM, CYH









 X線回折 / 使用した結晶の数: 1
X線回折 / 使用した結晶の数: 1  試料調製
試料調製 シンクロトロン / サイト:
シンクロトロン / サイト:  APS
APS  / ビームライン: 14-BM-C / 波長: 0.9 Å
/ ビームライン: 14-BM-C / 波長: 0.9 Å 解析
解析 分子置換
分子置換 ムービー
ムービー コントローラー
コントローラー














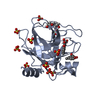
 PDBj
PDBj









