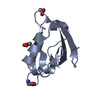
Entry Database : PDB / ID : 5mz7Title Crystal Structure of the third PDZ domain from the synaptic protein PSD-95 with incorporated Azidohomoalanine Disks large homolog 4 Keywords / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Rattus norvegicus (Norway rat)Method / / / Resolution : 1.53 Å Authors Kudlinzki, D. / Lehner, F. / Witt, K. / Linhard, V.L. / Silvers, R. / Schwalbe, H. Journal : Chembiochem / Year : 2017Title : Impact of Azidohomoalanine Incorporation on Protein Structure and Ligand Binding.Authors : Lehner, F. / Kudlinzki, D. / Richter, C. / Muller-Werkmeister, H.M. / Eberl, K.B. / Bredenbeck, J. / Schwalbe, H. / Silvers, R. History Deposition Jan 31, 2017 Deposition site / Processing site Revision 1.0 Oct 4, 2017 Provider / Type Revision 1.1 Oct 11, 2017 Group / Category / citation_author / Item / _citation.titleRevision 1.2 Nov 15, 2017 Group / Category / citation_author / Item / _citation_author.nameRevision 1.3 Dec 13, 2017 Group / Category Item / _citation.page_first / _citation.page_lastRevision 2.0 Apr 24, 2019 Group / Polymer sequence / Category / pdbx_seq_map_depositor_infoItem / _pdbx_seq_map_depositor_info.one_letter_codeRevision 2.1 Jun 12, 2019 Group / Structure summaryCategory / database_PDB_rev / database_PDB_rev_recordItem Revision 2.2 Oct 16, 2019 Group / Category / reflns_shell / Item / _reflns_shell.pdbx_CC_halfRevision 2.3 Jan 17, 2024 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Refinement description Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_conn Item / _database_2.pdbx_database_accession / _struct_conn.pdbx_leaving_atom_flag
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.53 Å
MOLECULAR REPLACEMENT / Resolution: 1.53 Å  Authors
Authors Citation
Citation Journal: Chembiochem / Year: 2017
Journal: Chembiochem / Year: 2017 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5mz7.cif.gz
5mz7.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5mz7.ent.gz
pdb5mz7.ent.gz PDB format
PDB format 5mz7.json.gz
5mz7.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/mz/5mz7
https://data.pdbj.org/pub/pdb/validation_reports/mz/5mz7 ftp://data.pdbj.org/pub/pdb/validation_reports/mz/5mz7
ftp://data.pdbj.org/pub/pdb/validation_reports/mz/5mz7

 Links
Links Assembly
Assembly




 Components
Components

 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SLS
SLS  / Beamline: X06DA / Wavelength: 1.00004 Å
/ Beamline: X06DA / Wavelength: 1.00004 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller











 PDBj
PDBj




