| Entry | Database: PDB / ID: 5j42
|
|---|
| Title | Crystal structure of m2hTDP2-CAT in complex with a small molecule inhibitor |
|---|
 Components Components | Tyrosyl-DNA phosphodiesterase 2 |
|---|
 Keywords Keywords | HYDROLASE / tyrosyl DNA phosphodiesterase 2 catalytic domain |
|---|
| Function / homology |  Function and homology information Function and homology information
tyrosyl-RNA phosphodiesterase activity / 5'-tyrosyl-DNA phosphodiesterase activity / Nonhomologous End-Joining (NHEJ) / Hydrolases; Acting on ester bonds; Phosphoric-diester hydrolases / phosphoric diester hydrolase activity / aggresome / neuron development / PML body / double-strand break repair / single-stranded DNA binding ...tyrosyl-RNA phosphodiesterase activity / 5'-tyrosyl-DNA phosphodiesterase activity / Nonhomologous End-Joining (NHEJ) / Hydrolases; Acting on ester bonds; Phosphoric-diester hydrolases / phosphoric diester hydrolase activity / aggresome / neuron development / PML body / double-strand break repair / single-stranded DNA binding / manganese ion binding / endonuclease activity / nucleolus / magnesium ion binding / cytoplasmSimilarity search - Function : / Deoxyribonuclease I; Chain A / Endonuclease/exonuclease/phosphatase / Endonuclease/exonuclease/phosphatase / Endonuclease/Exonuclease/phosphatase family / Endonuclease/exonuclease/phosphatase superfamily / 4-Layer Sandwich / Alpha BetaSimilarity search - Domain/homology |
|---|
| Biological species |   Mus musculus (house mouse) Mus musculus (house mouse) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.7 Å MOLECULAR REPLACEMENT / Resolution: 1.7 Å |
|---|
 Authors Authors | Hornyak, P. / Pearl, L.H. / Caldecott, K.W. / Oliver, A.W. |
|---|
| Funding support |  United Kingdom, 3items United Kingdom, 3items | Organization | Grant number | Country |
|---|
| Cancer Research UK | C302/A14532 |  United Kingdom United Kingdom | | Cancer Research UK | C6563/A16771 |  United Kingdom United Kingdom | | Cancer Research UK | C480/A11411 |  United Kingdom United Kingdom |
|
|---|
 Citation Citation |  Journal: Biochem.J. / Year: 2016 Journal: Biochem.J. / Year: 2016
Title: Mode of action of DNA-competitive small molecule inhibitors of tyrosyl DNA phosphodiesterase 2.
Authors: Hornyak, P. / Askwith, T. / Walker, S. / Komulainen, E. / Paradowski, M. / Pennicott, L.E. / Bartlett, E.J. / Brissett, N.C. / Raoof, A. / Watson, M. / Jordan, A.M. / Ogilvie, D.J. / Ward, S. ...Authors: Hornyak, P. / Askwith, T. / Walker, S. / Komulainen, E. / Paradowski, M. / Pennicott, L.E. / Bartlett, E.J. / Brissett, N.C. / Raoof, A. / Watson, M. / Jordan, A.M. / Ogilvie, D.J. / Ward, S.E. / Atack, J.R. / Pearl, L.H. / Caldecott, K.W. / Oliver, A.W. |
|---|
| History | | Deposition | Mar 31, 2016 | Deposition site: RCSB / Processing site: PDBE |
|---|
| Revision 1.0 | May 4, 2016 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Jul 6, 2016 | Group: Database references |
|---|
| Revision 1.2 | Jan 24, 2018 | Group: Source and taxonomy / Category: entity_src_gen / Item: _entity_src_gen.pdbx_host_org_ncbi_taxonomy_id |
|---|
| Revision 1.3 | Oct 16, 2019 | Group: Data collection / Category: reflns / reflns_shell / Item: _reflns.pdbx_CC_half / _reflns_shell.pdbx_CC_half |
|---|
| Revision 1.4 | Jan 10, 2024 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / pdbx_struct_conn_angle / struct_conn
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id |
|---|
| Revision 1.5 | Oct 23, 2024 | Group: Structure summary / Category: pdbx_entry_details / pdbx_modification_feature |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.7 Å
MOLECULAR REPLACEMENT / Resolution: 1.7 Å  Authors
Authors United Kingdom, 3items
United Kingdom, 3items  Citation
Citation Journal: Biochem.J. / Year: 2016
Journal: Biochem.J. / Year: 2016 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5j42.cif.gz
5j42.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5j42.ent.gz
pdb5j42.ent.gz PDB format
PDB format 5j42.json.gz
5j42.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/j4/5j42
https://data.pdbj.org/pub/pdb/validation_reports/j4/5j42 ftp://data.pdbj.org/pub/pdb/validation_reports/j4/5j42
ftp://data.pdbj.org/pub/pdb/validation_reports/j4/5j42



 Links
Links Assembly
Assembly
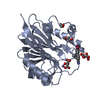

 Components
Components

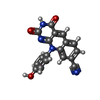










 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  Diamond
Diamond  / Beamline: I04 / Wavelength: 0.9795 Å
/ Beamline: I04 / Wavelength: 0.9795 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj






