[English] 日本語
 Yorodumi
Yorodumi- PDB-5j3z: Crystal structure of m2hTDP2-CAT in complex with a small molecule... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5j3z | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of m2hTDP2-CAT in complex with a small molecule inhibitor | ||||||||||||
 Components Components | Tyrosyl-DNA phosphodiesterase 2 | ||||||||||||
 Keywords Keywords | HYDROLASE / tyrosyl DNA phosphodiesterase 2 catalytic domain | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationtyrosyl-RNA phosphodiesterase activity / 5'-tyrosyl-DNA phosphodiesterase activity / Nonhomologous End-Joining (NHEJ) / Hydrolases; Acting on ester bonds; Phosphoric-diester hydrolases / phosphoric diester hydrolase activity / aggresome / neuron development / PML body / double-strand break repair / manganese ion binding ...tyrosyl-RNA phosphodiesterase activity / 5'-tyrosyl-DNA phosphodiesterase activity / Nonhomologous End-Joining (NHEJ) / Hydrolases; Acting on ester bonds; Phosphoric-diester hydrolases / phosphoric diester hydrolase activity / aggresome / neuron development / PML body / double-strand break repair / manganese ion binding / single-stranded DNA binding / endonuclease activity / nucleolus / magnesium ion binding / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.8 Å MOLECULAR REPLACEMENT / Resolution: 1.8 Å | ||||||||||||
 Authors Authors | Hornyak, P. / Pearl, L.H. / Caldecott, K.W. / Oliver, A.W. | ||||||||||||
| Funding support |  United Kingdom, 3items United Kingdom, 3items
| ||||||||||||
 Citation Citation |  Journal: Biochem.J. / Year: 2016 Journal: Biochem.J. / Year: 2016Title: Mode of action of DNA-competitive small molecule inhibitors of tyrosyl DNA phosphodiesterase 2. Authors: Hornyak, P. / Askwith, T. / Walker, S. / Komulainen, E. / Paradowski, M. / Pennicott, L.E. / Bartlett, E.J. / Brissett, N.C. / Raoof, A. / Watson, M. / Jordan, A.M. / Ogilvie, D.J. / Ward, S. ...Authors: Hornyak, P. / Askwith, T. / Walker, S. / Komulainen, E. / Paradowski, M. / Pennicott, L.E. / Bartlett, E.J. / Brissett, N.C. / Raoof, A. / Watson, M. / Jordan, A.M. / Ogilvie, D.J. / Ward, S.E. / Atack, J.R. / Pearl, L.H. / Caldecott, K.W. / Oliver, A.W. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5j3z.cif.gz 5j3z.cif.gz | 230.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5j3z.ent.gz pdb5j3z.ent.gz | 183.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5j3z.json.gz 5j3z.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/j3/5j3z https://data.pdbj.org/pub/pdb/validation_reports/j3/5j3z ftp://data.pdbj.org/pub/pdb/validation_reports/j3/5j3z ftp://data.pdbj.org/pub/pdb/validation_reports/j3/5j3z | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5j3pC  5j3sC  5j42C  4gyzS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 28776.164 Da / Num. of mol.: 2 / Fragment: UNP residues 118-370 / Mutation: E242G, Q278R, Y321C Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: Q9JJX7, Hydrolases; Acting on ester bonds; Phosphoric-diester hydrolases |
|---|
-Non-polymers , 6 types, 633 molecules 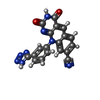










| #2: Chemical | | #3: Chemical | #4: Chemical | ChemComp-GOL / #5: Chemical | ChemComp-EDO / #6: Chemical | ChemComp-ACT / #7: Water | ChemComp-HOH / | |
|---|
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.5 Å3/Da / Density % sol: 50.79 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop Details: 0.1M Bis-Tris propane pH 7.5, 0.2 M sodium citrate, 20% w/v PEG3350, 0.3% v/v DMSO |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I04 / Wavelength: 0.9795 Å / Beamline: I04 / Wavelength: 0.9795 Å |
| Detector | Type: DECTRIS PILATUS 6M-F / Detector: PIXEL / Date: Oct 12, 2013 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9795 Å / Relative weight: 1 |
| Reflection | Resolution: 1.8→42.8 Å / Num. obs: 51393 / % possible obs: 98 % / Redundancy: 2.9 % / Biso Wilson estimate: 21.69 Å2 / CC1/2: 0.87 / Rmerge(I) obs: 0.051 / Net I/σ(I): 9.7 |
| Reflection shell | Resolution: 1.8→1.84 Å / Redundancy: 2.1 % / Rmerge(I) obs: 0.166 / Mean I/σ(I) obs: 2 / % possible all: 79.7 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4GYZ Resolution: 1.8→39.81 Å / Cor.coef. Fo:Fc: 0.9516 / Cor.coef. Fo:Fc free: 0.9338 / SU R Cruickshank DPI: 0.12 / Cross valid method: THROUGHOUT / σ(F): 0 / SU R Blow DPI: 0.132 / SU Rfree Blow DPI: 0.117 / SU Rfree Cruickshank DPI: 0.111
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 26.43 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.221 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.8→39.81 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.8→1.85 Å / Total num. of bins used: 20
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj






