[English] 日本語
 Yorodumi
Yorodumi- PDB-5ieg: Murine endoplasmic reticulum alpha-glucosidase II with N-9'-metho... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5ieg | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Murine endoplasmic reticulum alpha-glucosidase II with N-9'-methoxynonyl-1-deoxynojirimycin. | ||||||||||||
 Components Components |
| ||||||||||||
 Keywords Keywords | HYDROLASE / Enzyme Glycosyl hydrolase GH31 Quality control exoglycosidase / MON-DNJ | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationglucan 1,3-alpha-glucosidase activity / Glc2Man9GlcNAc2 oligosaccharide glucosidase activity / mannosyl-oligosaccharide alpha-1,3-glucosidase / glucosidase II complex / glucosidase activity / nitrogen cycle metabolic process / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / Post-translational protein phosphorylation / N-glycan processing / liver development ...glucan 1,3-alpha-glucosidase activity / Glc2Man9GlcNAc2 oligosaccharide glucosidase activity / mannosyl-oligosaccharide alpha-1,3-glucosidase / glucosidase II complex / glucosidase activity / nitrogen cycle metabolic process / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / Post-translational protein phosphorylation / N-glycan processing / liver development / melanosome / negative regulation of neuron projection development / carbohydrate binding / carbohydrate metabolic process / in utero embryonic development / intracellular membrane-bounded organelle / calcium ion binding / protein-containing complex binding / endoplasmic reticulum / Golgi apparatus / RNA binding Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  FOURIER SYNTHESIS / Resolution: 1.822 Å FOURIER SYNTHESIS / Resolution: 1.822 Å | ||||||||||||
 Authors Authors | Caputo, A.T. / Roversi, P. / Alonzi, D.S. / Kiappes, J.L. / Zitzmann, N. | ||||||||||||
| Funding support |  United Kingdom, 1items United Kingdom, 1items
| ||||||||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2016 Journal: Proc.Natl.Acad.Sci.USA / Year: 2016Title: Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals. Authors: Caputo, A.T. / Alonzi, D.S. / Marti, L. / Reca, I.B. / Kiappes, J.L. / Struwe, W.B. / Cross, A. / Basu, S. / Lowe, E.D. / Darlot, B. / Santino, A. / Roversi, P. / Zitzmann, N. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5ieg.cif.gz 5ieg.cif.gz | 231 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5ieg.ent.gz pdb5ieg.ent.gz | 175.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5ieg.json.gz 5ieg.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ie/5ieg https://data.pdbj.org/pub/pdb/validation_reports/ie/5ieg ftp://data.pdbj.org/pub/pdb/validation_reports/ie/5ieg ftp://data.pdbj.org/pub/pdb/validation_reports/ie/5ieg | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5f0eSC  5h9oC  5hjoC  5hjrC  5iedC  5ieeC  5iefC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 2 types, 2 molecules AB
| #1: Protein | Mass: 103818.969 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: This chain contains all of the residues from the mature Q8BHN3 isoform 2 (i.e. no signal peptide and starts at residue 33) but has been trypsinised so that there are two gaps in the sequence ...Details: This chain contains all of the residues from the mature Q8BHN3 isoform 2 (i.e. no signal peptide and starts at residue 33) but has been trypsinised so that there are two gaps in the sequence between: 186-243 and 351-369 (inclusive) Source: (gene. exp.)   Homo sapiens (human) / References: UniProt: Q8BHN3, EC: 3.2.1.84 Homo sapiens (human) / References: UniProt: Q8BHN3, EC: 3.2.1.84 |
|---|---|
| #2: Protein | Mass: 9568.298 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: This chain contains all of the residues from the mature O08795 but has been trypsinised so that there all that was crystallised are residues: 30-117 Source: (gene. exp.)   Homo sapiens (human) / References: UniProt: O08795 Homo sapiens (human) / References: UniProt: O08795 |
-Sugars , 1 types, 1 molecules
| #3: Polysaccharide | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source |
|---|
-Non-polymers , 7 types, 506 molecules 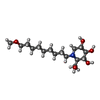












| #4: Chemical | ChemComp-6A9 / | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| #5: Chemical | ChemComp-FMT / #6: Chemical | ChemComp-P6G / #7: Chemical | ChemComp-DMS / | #8: Chemical | #9: Chemical | #10: Water | ChemComp-HOH / | |
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.65 Å3/Da / Density % sol: 53.54 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop Details: 21% v/v ethylene glycol, 11% w/v PEG 8000 (from the Morpheus Precipitant Mix 2), 50 mM Morpheus carboxylic acids mix, 100 mM Morpheus buffer system 1 pH 6.25 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I02 / Wavelength: 1.07205 Å / Beamline: I02 / Wavelength: 1.07205 Å |
| Detector | Type: DECTRIS PILATUS 6M-F / Detector: PIXEL / Date: Feb 11, 2016 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.07205 Å / Relative weight: 1 |
| Reflection | Resolution: 1.822→104.321 Å / Num. obs: 102902 / % possible obs: 100 % / Redundancy: 6.7 % / Biso Wilson estimate: 24 Å2 / CC1/2: 0.998 / Rsym value: 0.129 / Net I/σ(I): 12.1 |
| Reflection shell | Resolution: 1.822→1.828 Å / Redundancy: 6.9 % / Mean I/σ(I) obs: 2.1 / % possible all: 97.5 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  FOURIER SYNTHESIS FOURIER SYNTHESISStarting model: 5F0E Resolution: 1.822→89.38 Å / Cor.coef. Fo:Fc: 0.953 / Cor.coef. Fo:Fc free: 0.94 / SU R Cruickshank DPI: 0.111 / Cross valid method: THROUGHOUT / σ(F): 0 / SU R Blow DPI: 0.112 / SU Rfree Blow DPI: 0.101 / SU Rfree Cruickshank DPI: 0.101
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 26.13 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Resolution: 1.822→89.38 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.822→1.87 Å / Rfactor Rfree error: 0
|
 Movie
Movie Controller
Controller












 PDBj
PDBj












