Entry Database : PDB / ID : 5hqiTitle Insulin with proline analog HzP at position B28 in the T2 state Insulin A-Chain Insulin B-Chain Keywords / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / / / Resolution : 0.97 Å Authors Lieblich, S.A. / Fang, K.Y. / Cahn, J.K.B. / Tirrell, D.A. Funding support Organization Grant number Country Novo Nordisk Foundation
Journal : J. Am. Chem. Soc. / Year : 2017Title : 4S-Hydroxylation of Insulin at ProB28 Accelerates Hexamer Dissociation and Delays Fibrillation.Authors : Lieblich, S.A. / Fang, K.Y. / Cahn, J.K.B. / Rawson, J. / LeBon, J. / Ku, H.T. / Tirrell, D.A. History Deposition Jan 21, 2016 Deposition site / Processing site Revision 1.0 Jan 25, 2017 Provider / Type Revision 1.1 Jul 12, 2017 Group / Category / citation_authorItem _citation.country / _citation.journal_abbrev ... _citation.country / _citation.journal_abbrev / _citation.journal_id_ASTM / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.journal_volume / _citation.page_first / _citation.page_last / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year Revision 1.2 Jan 15, 2020 Group / Category / Item Revision 1.3 Sep 27, 2023 Group / Database references / Refinement descriptionCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / diffrn_radiation_wavelength / pdbx_initial_refinement_model Item / _database_2.pdbx_database_accessionRevision 1.4 Nov 15, 2023 Group / Category / chem_comp_bond / Item / _chem_comp_bond.atom_id_2
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT /
MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 0.97 Å
molecular replacement / Resolution: 0.97 Å  Authors
Authors Denmark, 1items
Denmark, 1items  Citation
Citation Journal: J. Am. Chem. Soc. / Year: 2017
Journal: J. Am. Chem. Soc. / Year: 2017 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5hqi.cif.gz
5hqi.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5hqi.ent.gz
pdb5hqi.ent.gz PDB format
PDB format 5hqi.json.gz
5hqi.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/hq/5hqi
https://data.pdbj.org/pub/pdb/validation_reports/hq/5hqi ftp://data.pdbj.org/pub/pdb/validation_reports/hq/5hqi
ftp://data.pdbj.org/pub/pdb/validation_reports/hq/5hqi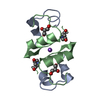


 Links
Links Assembly
Assembly

 Components
Components Homo sapiens (human) / Gene: INS / Plasmid: pQE80L / Production host:
Homo sapiens (human) / Gene: INS / Plasmid: pQE80L / Production host: 
 Homo sapiens (human) / Gene: INS / Plasmid: pQE80L / Production host:
Homo sapiens (human) / Gene: INS / Plasmid: pQE80L / Production host: 
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SSRL
SSRL  / Beamline: BL12-2 / Wavelength: 0.8265 Å
/ Beamline: BL12-2 / Wavelength: 0.8265 Å molecular replacement
molecular replacement Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller













 PDBj
PDBj

















