[English] 日本語
 Yorodumi
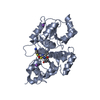
Yorodumi- PDB-5c1e: Crystal Structure of the Pectin Methylesterase from Aspergillus n... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5c1e | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal Structure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) | ||||||
 Components Components | Pectinesterase | ||||||
 Keywords Keywords | HYDROLASE / parallel beta helix / pectin methylesterase | ||||||
| Function / homology |  Function and homology information Function and homology informationpectinesterase / pectinesterase activity / cell wall modification / pectin catabolic process / extracellular region Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.75 Å molecular replacement / Resolution: 1.75 Å | ||||||
| Model details | Protein deglycosylated with EndoHf to leave an Asn84 N-linked N-acetylglycosamine stub | ||||||
 Authors Authors | Jameson, G.B. / Williams, M.A.K. / Loo, T.S. / Kent, L.M. / Melton, L.D. / Mercadante, D. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2016 Journal: J.Biol.Chem. / Year: 2016Title: Structure and Properties of a Non-processive, Salt-requiring, and Acidophilic Pectin Methylesterase from Aspergillus niger Provide Insights into the Key Determinants of Processivity Control. Authors: Kent, L.M. / Loo, T.S. / Melton, L.D. / Mercadante, D. / Williams, M.A. / Jameson, G.B. #1:  Journal: PLoS ONE / Year: 2014 Journal: PLoS ONE / Year: 2014Title: Processive pectin methylesterases: the role of electrostatic potential, breathing motions and bond cleavage in the rectification of Brownian motions Authors: Mercadante, D. / Melton, L.D. / Jameson, G.B. / Williams, M.A.K. #2:  Journal: Biophys J / Year: 2013 Journal: Biophys J / Year: 2013Title: Substrate Dynamics in Enzyme Action: Rotations of Monosaccharide Subunits in the Binding Groove are Essential for Pectin Methylesterase Processivity Authors: Mercadante, D. / Melton, L.D. / Jameson, G.B. / Williams, M.A.K. / De Simone, A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5c1e.cif.gz 5c1e.cif.gz | 135.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5c1e.ent.gz pdb5c1e.ent.gz | 104.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5c1e.json.gz 5c1e.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/c1/5c1e https://data.pdbj.org/pub/pdb/validation_reports/c1/5c1e ftp://data.pdbj.org/pub/pdb/validation_reports/c1/5c1e ftp://data.pdbj.org/pub/pdb/validation_reports/c1/5c1e | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5c1cC  1xg2S C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||||||||
| Unit cell |
| |||||||||||||||
| Components on special symmetry positions |
| |||||||||||||||
| Details | Monomer according to Gel filtration |
- Components
Components
-Protein / Sugars , 2 types, 2 molecules A

| #1: Protein | Mass: 31792.287 Da / Num. of mol.: 1 / Fragment: N-terminal truncated exported protein Source method: isolated from a genetically manipulated source Details: After purification, protein was deglycosylated with PNGaseF Source: (gene. exp.)  Strain: van Tieghem, anamorph / Gene: ASPNIDRAFT_214857 / Plasmid: pYES2 / Production host:  |
|---|---|
| #2: Sugar | ChemComp-NAG / |
-Non-polymers , 5 types, 335 molecules 








| #3: Chemical | ChemComp-SO4 / #4: Chemical | ChemComp-CL / | #5: Chemical | ChemComp-GOL / #6: Chemical | ChemComp-ACT / | #7: Water | ChemComp-HOH / | |
|---|
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.96 Å3/Da / Density % sol: 58.4 % |
|---|---|
| Crystal grow | Temperature: 294 K / Method: vapor diffusion, hanging drop / pH: 3.6 Details: Protein (6.5 mg/mL) in 50-100 mM acetate buffer mixed 1:1 with 1.9 M ammonium sulfate, 100 mM sodium acetate, pH 3.6 |
-Data collection
| Diffraction | Mean temperature: 123 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU MICROMAX-007 / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU MICROMAX-007 / Wavelength: 1.5418 Å |
| Detector | Type: RIGAKU RAXIS IV++ / Detector: IMAGE PLATE / Date: Nov 4, 2013 / Details: AXCo PX70 QUARTZ GLASS CAPILLARY OPTIC |
| Radiation | Monochromator: AXCo PX70 QUARTZ GLASS CAPILLARY OPTIC / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.75→36.16 Å / Num. all: 36656 / Num. obs: 36656 / % possible obs: 95.6 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 3.94 % / Biso Wilson estimate: 19 Å2 / Rmerge(I) obs: 0.084 / Net I/av σ(I): 8.3 / Net I/σ(I): 4.5 |
| Reflection shell | Resolution: 1.75→1.8 Å / Redundancy: 3.71 % / Rmerge(I) obs: 0.366 / Mean I/σ(I) obs: 3.1 / % possible all: 89.6 |
-Phasing
| Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1xg2 Resolution: 1.75→36.16 Å / Cor.coef. Fo:Fc: 0.964 / Cor.coef. Fo:Fc free: 0.948 / WRfactor Rfree: 0.1975 / WRfactor Rwork: 0.1688 / FOM work R set: 0.8722 / SU B: 4.407 / SU ML: 0.071 / SU R Cruickshank DPI: 0.1049 / SU Rfree: 0.1022 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.105 / ESU R Free: 0.102 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS U VALUES : WITH TLS ADDED
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 61.96 Å2 / Biso mean: 20.727 Å2 / Biso min: 12.39 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.75→36.16 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.75→1.795 Å / Total num. of bins used: 20
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: -19.2157 Å / Origin y: 25.7632 Å / Origin z: 12.2763 Å
|
 Movie
Movie Controller
Controller











 PDBj
PDBj




