+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1boh | ||||||
|---|---|---|---|---|---|---|---|
| Title | SULFUR-SUBSTITUTED RHODANESE (ORTHORHOMBIC FORM) | ||||||
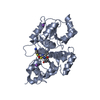
 Components Components | RHODANESE | ||||||
 Keywords Keywords | TRANSFERASE / RHODANESE / SULFURTRANSFERASE | ||||||
| Function / homology |  Function and homology information Function and homology informationrRNA transport / 3-mercaptopyruvate sulfurtransferase activity / thiosulfate sulfurtransferase / thiosulfate-cyanide sulfurtransferase activity / rRNA import into mitochondrion / 5S rRNA binding / mitochondrial matrix / mitochondrion Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.3 Å MOLECULAR REPLACEMENT / Resolution: 2.3 Å | ||||||
 Authors Authors | Gliubich, F. / Berni, R. / Cianci, M. / Trevino, R.J. / Horowitz, P.M. / Zanotti, G. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 1999 Journal: J.Biol.Chem. / Year: 1999Title: NH2-terminal sequence truncation decreases the stability of bovine rhodanese, minimally perturbs its crystal structure, and enhances interaction with GroEL under native conditions. Authors: Trevino, R.J. / Gliubich, F. / Berni, R. / Cianci, M. / Chirgwin, J.M. / Zanotti, G. / Horowitz, P.M. #1:  Journal: Acta Crystallogr.,Sect.D / Year: 1998 Journal: Acta Crystallogr.,Sect.D / Year: 1998Title: Structure of Sulfur-Substituted Rhodanese at 1.36 A Resolution Authors: Gliubich, F. / Berni, R. / Colapietro, M. / Barba, L. / Zanotti, G. #2:  Journal: J.Biol.Chem. / Year: 1996 Journal: J.Biol.Chem. / Year: 1996Title: Active Site Structural Features for Chemically Modified Forms of Rhodanese Authors: Gliubich, F. / Gazerro, M. / Zanotti, G. / Delbono, S. / Bombieri, G. / Berni, R. #3:  Journal: J.Mol.Biol. / Year: 1979 Journal: J.Mol.Biol. / Year: 1979Title: The Structure of Bovine Liver Rhodanese. II. The Active Site in the Sulfur-Substituted and the Sulfur-Free Enzyme Authors: Ploegman, J.H. / Drent, G. / Kalk, K.H. / Hol, W.G. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1boh.cif.gz 1boh.cif.gz | 71.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1boh.ent.gz pdb1boh.ent.gz | 52.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1boh.json.gz 1boh.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bo/1boh https://data.pdbj.org/pub/pdb/validation_reports/bo/1boh ftp://data.pdbj.org/pub/pdb/validation_reports/bo/1boh ftp://data.pdbj.org/pub/pdb/validation_reports/bo/1boh | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1boiC  1rhsS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 33240.668 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: SULFUR SUBSTITUTED AT RESIDUE CYS 247 / Source: (gene. exp.)   |
|---|---|
| #2: Water | ChemComp-HOH / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 2 X-RAY DIFFRACTION / Number of used crystals: 2 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.9 Å3/Da / Density % sol: 37 % | ||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 7.3 / Details: pH 7.3 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 4 ℃ / Method: vapor diffusion, sitting drop | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 297 K |
|---|---|
| Diffraction source | Wavelength: 1.5418 |
| Detector | Type: SIEMENS / Detector: AREA DETECTOR / Date: Oct 1, 1997 |
| Radiation | Monochromator: GRAPHITE(002) / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.3→55 Å / Num. obs: 10362 / % possible obs: 78.8 % / Observed criterion σ(I): 0 / Redundancy: 5.9 % / Rmerge(I) obs: 0.11 / Rsym value: 0.11 |
| Reflection shell | Resolution: 2.3→2.42 Å / Redundancy: 1.3 % / Rmerge(I) obs: 0.34 / Rsym value: 0.24 / % possible all: 13.4 |
| Reflection | *PLUS Num. measured all: 60362 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1RHS Resolution: 2.3→55 Å / Data cutoff high absF: 1000000 / Data cutoff low absF: 0.0001 / Cross valid method: FREE R-VALUE / σ(F): 0
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.3→55 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file | Serial no: 1 / Param file: PARAM19X.PRO / Topol file: TOPH19X.PRO | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.83 / Classification: refinement X-PLOR / Version: 3.83 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller












 PDBj
PDBj
