[English] 日本語
 Yorodumi
Yorodumi- PDB-5bna: THE PRIMARY MODE OF BINDING OF CISPLATIN TO A B-DNA DODECAMER: C-... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5bna | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | THE PRIMARY MODE OF BINDING OF CISPLATIN TO A B-DNA DODECAMER: C-G-C-G-A-A-T-T-C-G-C-G | ||||||||||||||||||
 Components Components | DNA (5'-D(* Keywords KeywordsDNA / B-DNA / DOUBLE HELIX / COMPLEXED WITH DRUG / MODIFIED | Function / homology | PLATINUM TRIAMINE ION / DNA / DNA (> 10) |  Function and homology information Function and homology informationMethod |  X-RAY DIFFRACTION / Resolution: 2.6 Å X-RAY DIFFRACTION / Resolution: 2.6 Å  Authors AuthorsWing, R.M. / Pjura, P. / Drew, H.R. / Dickerson, R.E. |  Citation Citation Journal: EMBO J. / Year: 1984 Journal: EMBO J. / Year: 1984Title: The primary mode of binding of cisplatin to a B-DNA dodecamer: C-G-C-G-A-A-T-T-C-G-C-G Authors: Wing, R.M. / Pjura, P. / Drew, H.R. / Dickerson, R.E. #1:  Journal: Proc.Natl.Acad.Sci.USA / Year: 1983 Journal: Proc.Natl.Acad.Sci.USA / Year: 1983Title: A Random-Walk Model for Helix Bending in B-DNA Authors: Dickerson, R.E. / Kopka, M.L. / Pjura, P. #2:  Journal: Cold Spring Harbor Symp.Quant.Biol. / Year: 1983 Journal: Cold Spring Harbor Symp.Quant.Biol. / Year: 1983Title: Helix Geometry and Hydration in A-DNA, B-DNA and Z-DNA Authors: Dickerson, R.E. / Drew, H.R. / Conner, B.N. / Kopka, M.L. / Pjura, P.E. #3:  Journal: Biological Macromolecules and Assemblies / Year: 1985 Journal: Biological Macromolecules and Assemblies / Year: 1985Title: Base Sequence, Helix Geometry, Hydration and Helix Stability in B-DNA Authors: Dickerson, R.E. / Kopka, M.L. / Pjura, P. #4:  Journal: Nature / Year: 1980 Journal: Nature / Year: 1980Title: Crystal Structure Analysis of a Complete Turn of B-DNA Authors: Wing, R. / Drew, H.R. / Takano, T. / Broka, C. / Tanaka, S. / Itakura, K. / Dickerson, R.E. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5bna.cif.gz 5bna.cif.gz | 28.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5bna.ent.gz pdb5bna.ent.gz | 18.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5bna.json.gz 5bna.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bn/5bna https://data.pdbj.org/pub/pdb/validation_reports/bn/5bna ftp://data.pdbj.org/pub/pdb/validation_reports/bn/5bna ftp://data.pdbj.org/pub/pdb/validation_reports/bn/5bna | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 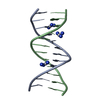
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Atom site foot note | 1: THE ANISOTROPIC TEMPERATURE FACTORS FOR THE PLATINUM ATOMS ARE AS FOLLOWS ATOM B11 B22 B33 B12 B13 B23 PT1 PTN 25 0.01510 0.00438 0.00138 0.00214 0.00056 0.00026 PT1 PTN 26 0.01545 0.00544 0.00131- ...1: THE ANISOTROPIC TEMPERATURE FACTORS FOR THE PLATINUM ATOMS ARE AS FOLLOWS ATOM B11 B22 B33 B12 B13 B23 PT1 PTN 25 0.01510 0.00438 0.00138 0.00214 0.00056 0.00026 PT1 PTN 26 0.01545 0.00544 0.00131-0.00462-0.00072 0.00017 PT1 PTN 27 0.00729 0.00382 0.00120-0.00092-0.00060 0.00039 2: TEMPERATURE FACTORS WHICH WERE CALCULATED TO BE NON-POSITIVE-DEFINITE WERE ARBITRARILY SET TO 0.0 IN THE REFINEMENT PROCEDURE. |
- Components
Components
| #1: DNA chain | Mass: 3663.392 Da / Num. of mol.: 2 / Source method: obtained synthetically #2: Chemical | #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.18 Å3/Da / Density % sol: 43.49 % | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion / Details: VAPOR DIFFUSION, temperature 277.00K | |||||||||||||||||||||||||
| Components of the solutions |
| |||||||||||||||||||||||||
| Crystal grow | *PLUS Details: Wing, R., (1980) Nature, 287, 755. | |||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Reflection | Highest resolution: 2.6 Å / Num. obs: 2088 |
|---|
- Processing
Processing
| Software | Name: JACK-LEVITT / Classification: refinement | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 2.6→8 Å / σ(F): 2 /
| ||||||||||||||||
| Refine Biso |
| ||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.6→8 Å
| ||||||||||||||||
| Refinement | *PLUS Highest resolution: 2.6 Å / Lowest resolution: 8 Å / Num. reflection all: 2012 / σ(F): 2 / Rfactor all: 0.166 / Rfactor obs: 0.112 | ||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller












 PDBj
PDBj









































