[English] 日本語
 Yorodumi
Yorodumi- PDB-5b8d: Crystal structure of a low occupancy fragment candidate (N-(4-Met... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5b8d | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of a low occupancy fragment candidate (N-(4-Methyl-1,3-thiazol-2-yl)propanamide) bound adjacent to the ubiquitin binding pocket of the HDAC6 zinc-finger domain | ||||||
 Components Components | Histone deacetylase 6 | ||||||
 Keywords Keywords | HYDROLASE / HISTONE DEACETYLASE / HDAC / HDAC6 / FRAGMENT SCREENING / STRUCTURAL GENOMICS CONSORTIUM / SGC / DIAMOND I04-1 XCHEM / PANDDA | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of hydrogen peroxide metabolic process / cellular response to topologically incorrect protein / polyubiquitinated misfolded protein transport / positive regulation of cellular response to oxidative stress / negative regulation of aggrephagy / positive regulation of cholangiocyte proliferation / response to misfolded protein / positive regulation of protein oligomerization / negative regulation of axon extension involved in axon guidance / type 2 mitophagy ...negative regulation of hydrogen peroxide metabolic process / cellular response to topologically incorrect protein / polyubiquitinated misfolded protein transport / positive regulation of cellular response to oxidative stress / negative regulation of aggrephagy / positive regulation of cholangiocyte proliferation / response to misfolded protein / positive regulation of protein oligomerization / negative regulation of axon extension involved in axon guidance / type 2 mitophagy / positive regulation of RIG-I signaling pathway / negative regulation of protein-containing complex disassembly / peroxidase inhibitor activity / erythrocyte enucleation / regulation of autophagy of mitochondrion / Cilium Assembly / tubulin deacetylation / protein-containing complex disassembly / regulation of establishment of protein localization / collateral sprouting / Transcriptional regulation by RUNX2 / tubulin deacetylase activity / negative regulation of microtubule depolymerization / lysosome localization / ATPase inhibitor activity / cilium disassembly / histone deacetylase activity, hydrolytic mechanism / misfolded protein binding / ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway / dendritic spine morphogenesis / positive regulation of type 2 mitophagy / protein deacetylation / aggresome assembly / positive regulation of dendrite morphogenesis / regulation of androgen receptor signaling pathway / regulation of mitochondrion organization / Transferases; Acyltransferases; Aminoacyltransferases / cellular response to misfolded protein / regulation of fat cell differentiation / protein lysine deacetylase activity / Hydrolases; Acting on carbon-nitrogen bonds, other than peptide bonds; In linear amides / histone deacetylase activity / aggresome / microtubule associated complex / cellular response to parathyroid hormone stimulus / positive regulation of intracellular estrogen receptor signaling pathway / response to dexamethasone / response to corticosterone / Notch-HLH transcription pathway / axonal transport of mitochondrion / negative regulation of gene expression, epigenetic / regulation of microtubule-based movement / RUNX2 regulates osteoblast differentiation / histone deacetylase complex / response to immobilization stress / cell leading edge / dynein complex binding / protein quality control for misfolded or incompletely synthesized proteins / positive regulation of epithelial cell migration / polyubiquitin modification-dependent protein binding / cilium assembly / regulation of macroautophagy / HSF1 activation / positive regulation of synaptic transmission, glutamatergic / alpha-tubulin binding / beta-tubulin binding / negative regulation of protein-containing complex assembly / negative regulation of proteolysis / multivesicular body / inclusion body / axon cytoplasm / antiviral innate immune response / epigenetic regulation of gene expression / actin filament organization / ubiquitin binding / transcription corepressor binding / response to amphetamine / intracellular protein transport / Hsp90 protein binding / regulation of protein stability / Late endosomal microautophagy / beta-catenin binding / caveola / regulation of autophagy / NOTCH1 Intracellular Domain Regulates Transcription / Constitutive Signaling by NOTCH1 PEST Domain Mutants / Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants / protein destabilization / tau protein binding / epidermal growth factor receptor signaling pathway / cellular response to hydrogen peroxide / histone deacetylase binding / protein polyubiquitination / Chaperone Mediated Autophagy / Aggrephagy / cellular response to heat / actin binding / microtubule binding / microtubule / perikaryon Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 1.05 Å SYNCHROTRON / Resolution: 1.05 Å | ||||||
 Authors Authors | Harding, R.J. / Tempel, W. / Collins, P. / Pearce, N. / Brandao-Neto, J. / Douangamath, A. / Ravichandran, M. / Schapira, M. / Bountra, C. / Edwards, A.M. ...Harding, R.J. / Tempel, W. / Collins, P. / Pearce, N. / Brandao-Neto, J. / Douangamath, A. / Ravichandran, M. / Schapira, M. / Bountra, C. / Edwards, A.M. / von Delft, F. / Santhakumar, V. / Arrowsmith, C.M. / Structural Genomics Consortium (SGC) | ||||||
 Citation Citation |  Journal: J. Med. Chem. / Year: 2017 Journal: J. Med. Chem. / Year: 2017Title: Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin. Authors: Harding, R.J. / Ferreira de Freitas, R. / Collins, P. / Franzoni, I. / Ravichandran, M. / Ouyang, H. / Juarez-Ornelas, K.A. / Lautens, M. / Schapira, M. / von Delft, F. / Santhakumar, V. / Arrowsmith, C.H. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5b8d.cif.gz 5b8d.cif.gz | 65.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5b8d.ent.gz pdb5b8d.ent.gz | 46.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5b8d.json.gz 5b8d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/b8/5b8d https://data.pdbj.org/pub/pdb/validation_reports/b8/5b8d ftp://data.pdbj.org/pub/pdb/validation_reports/b8/5b8d ftp://data.pdbj.org/pub/pdb/validation_reports/b8/5b8d | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5kh3C  5kh7C  5kh9C  5wpbC  3c5kS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 11932.607 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: HDAC6, KIAA0901, JM21 / Plasmid: pET28-lic / Production host: Homo sapiens (human) / Gene: HDAC6, KIAA0901, JM21 / Plasmid: pET28-lic / Production host:  |
|---|
-Non-polymers , 6 types, 122 molecules 

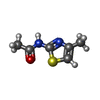








| #2: Chemical | | #3: Chemical | #4: Chemical | ChemComp-6T4 / ~{ | #5: Chemical | ChemComp-UNX / #6: Chemical | ChemComp-NA / | #7: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.17 Å3/Da / Density % sol: 43.26 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, sitting drop / pH: 4.6 Details: 2M Na-formate, 0.2M Na-acetate pH4.6, 5% ethylene glycol |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I04-1 / Wavelength: 0.92819 Å / Beamline: I04-1 / Wavelength: 0.92819 Å | ||||||||||||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS 6M-F / Detector: PIXEL / Date: Sep 30, 2015 | ||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.92819 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.05→34.8 Å / Num. obs: 47523 / % possible obs: 97.4 % / Redundancy: 5.4 % / CC1/2: 0.999 / Rmerge(I) obs: 0.058 / Rpim(I) all: 0.026 / Rrim(I) all: 0.064 / Net I/σ(I): 14.6 / Num. measured all: 257744 | ||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1 / Rejects: _
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Starting model: pdbid 3C5K Resolution: 1.05→34.8 Å / Cor.coef. Fo:Fc: 0.98 / Cor.coef. Fo:Fc free: 0.977 / SU B: 0.733 / SU ML: 0.016 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.023 / ESU R Free: 0.023 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: Users of this crystal structure: verify our intepretion of the electron density. Amplitudes and unmerged intensities are included with this deposition. Diffraction images will be deposited ...Details: Users of this crystal structure: verify our intepretion of the electron density. Amplitudes and unmerged intensities are included with this deposition. Diffraction images will be deposited in a public repository. Geometry restraints for the fragment candidate were prepared with GRADE. The methyl group of the fragment candidate was not resolved by electron density and was omitted from the model. Ambiguous difference density suggests some form of covalent modification of Cys1147.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 69.28 Å2 / Biso mean: 13.189 Å2 / Biso min: 6.88 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.05→34.8 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.052→1.08 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller










 PDBj
PDBj









