[English] 日本語
 Yorodumi
Yorodumi- PDB-5kh9: Crystal structure of a low occupancy fragment candidate (5-[(4-Is... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5kh9 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of a low occupancy fragment candidate (5-[(4-Isopropylphenyl)amino]-6-methyl-1,2,4-triazin-3(2H)-one) bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | ||||||
 Components Components | Histone deacetylase 6 | ||||||
 Keywords Keywords | HYDROLASE / HISTONE DEACETYLASE / HDAC / HDAC6 / FRAGMENT SCREENING / STRUCTURAL GENOMICS CONSORTIUM / SGC / DIAMOND I04-1 XCHEM / PANDDA | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of hydrogen peroxide metabolic process / cellular response to topologically incorrect protein / polyubiquitinated misfolded protein transport / positive regulation of cellular response to oxidative stress / negative regulation of aggrephagy / positive regulation of cholangiocyte proliferation / response to misfolded protein / positive regulation of protein oligomerization / negative regulation of axon extension involved in axon guidance / type 2 mitophagy ...negative regulation of hydrogen peroxide metabolic process / cellular response to topologically incorrect protein / polyubiquitinated misfolded protein transport / positive regulation of cellular response to oxidative stress / negative regulation of aggrephagy / positive regulation of cholangiocyte proliferation / response to misfolded protein / positive regulation of protein oligomerization / negative regulation of axon extension involved in axon guidance / type 2 mitophagy / positive regulation of RIG-I signaling pathway / negative regulation of protein-containing complex disassembly / peroxidase inhibitor activity / erythrocyte enucleation / regulation of autophagy of mitochondrion / Cilium Assembly / tubulin deacetylation / protein-containing complex disassembly / regulation of establishment of protein localization / collateral sprouting / Transcriptional regulation by RUNX2 / tubulin deacetylase activity / negative regulation of microtubule depolymerization / lysosome localization / ATPase inhibitor activity / cilium disassembly / histone deacetylase activity, hydrolytic mechanism / misfolded protein binding / ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway / dendritic spine morphogenesis / positive regulation of type 2 mitophagy / protein deacetylation / aggresome assembly / positive regulation of dendrite morphogenesis / regulation of androgen receptor signaling pathway / regulation of mitochondrion organization / Transferases; Acyltransferases; Aminoacyltransferases / cellular response to misfolded protein / regulation of fat cell differentiation / protein lysine deacetylase activity / Hydrolases; Acting on carbon-nitrogen bonds, other than peptide bonds; In linear amides / histone deacetylase activity / aggresome / microtubule associated complex / cellular response to parathyroid hormone stimulus / positive regulation of intracellular estrogen receptor signaling pathway / response to corticosterone / response to dexamethasone / Notch-HLH transcription pathway / axonal transport of mitochondrion / negative regulation of gene expression, epigenetic / regulation of microtubule-based movement / RUNX2 regulates osteoblast differentiation / histone deacetylase complex / response to immobilization stress / cell leading edge / dynein complex binding / protein quality control for misfolded or incompletely synthesized proteins / positive regulation of epithelial cell migration / cilium assembly / polyubiquitin modification-dependent protein binding / regulation of macroautophagy / HSF1 activation / alpha-tubulin binding / positive regulation of synaptic transmission, glutamatergic / beta-tubulin binding / negative regulation of protein-containing complex assembly / negative regulation of proteolysis / multivesicular body / inclusion body / axon cytoplasm / antiviral innate immune response / epigenetic regulation of gene expression / actin filament organization / ubiquitin binding / transcription corepressor binding / response to amphetamine / intracellular protein transport / Hsp90 protein binding / regulation of protein stability / Late endosomal microautophagy / beta-catenin binding / caveola / regulation of autophagy / NOTCH1 Intracellular Domain Regulates Transcription / protein destabilization / Constitutive Signaling by NOTCH1 PEST Domain Mutants / Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants / tau protein binding / epidermal growth factor receptor signaling pathway / histone deacetylase binding / cellular response to hydrogen peroxide / protein polyubiquitination / Chaperone Mediated Autophagy / Aggrephagy / cellular response to heat / actin binding / microtubule binding / microtubule / perikaryon Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 1.07 Å SYNCHROTRON / Resolution: 1.07 Å | ||||||
 Authors Authors | Harding, R.J. / Tempel, W. / Ravichandran, M. / Collins, P. / Pearce, N. / Brandao-Neto, J. / Douangamath, A. / Schapira, M. / Bountra, C. / Edwards, A.M. ...Harding, R.J. / Tempel, W. / Ravichandran, M. / Collins, P. / Pearce, N. / Brandao-Neto, J. / Douangamath, A. / Schapira, M. / Bountra, C. / Edwards, A.M. / von Delft, F. / Santhakumar, V. / Arrowsmith, C.M. / Structural Genomics Consortium (SGC) | ||||||
 Citation Citation |  Journal: J. Med. Chem. / Year: 2017 Journal: J. Med. Chem. / Year: 2017Title: Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin. Authors: Harding, R.J. / Ferreira de Freitas, R. / Collins, P. / Franzoni, I. / Ravichandran, M. / Ouyang, H. / Juarez-Ornelas, K.A. / Lautens, M. / Schapira, M. / von Delft, F. / Santhakumar, V. / Arrowsmith, C.H. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5kh9.cif.gz 5kh9.cif.gz | 65.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5kh9.ent.gz pdb5kh9.ent.gz | 45.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5kh9.json.gz 5kh9.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/kh/5kh9 https://data.pdbj.org/pub/pdb/validation_reports/kh/5kh9 ftp://data.pdbj.org/pub/pdb/validation_reports/kh/5kh9 ftp://data.pdbj.org/pub/pdb/validation_reports/kh/5kh9 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5b8dC  5kh3C  5kh7C  5wpbC  3c5kS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 11932.607 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: HDAC6, KIAA0901, JM21 / Plasmid: pET28-lic / Production host: Homo sapiens (human) / Gene: HDAC6, KIAA0901, JM21 / Plasmid: pET28-lic / Production host:  |
|---|
-Non-polymers , 6 types, 118 molecules 

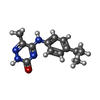








| #2: Chemical | | #3: Chemical | ChemComp-FMT / | #4: Chemical | ChemComp-6T5 / | #5: Chemical | ChemComp-UNX / #6: Chemical | ChemComp-NA / | #7: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.17 Å3/Da / Density % sol: 43.26 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, sitting drop / pH: 4.6 Details: 2M Na-formate, 0.2M Na-acetate pH4.6, 5% ethylene glycol |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I04-1 / Wavelength: 0.92819 Å / Beamline: I04-1 / Wavelength: 0.92819 Å | ||||||||||||||||||
| Detector | Type: DECTRIS PILATUS 6M-F / Detector: PIXEL / Date: Oct 2, 2015 | ||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||
| Radiation wavelength | Wavelength: 0.92819 Å / Relative weight: 1 | ||||||||||||||||||
| Reflection | Resolution: 1.07→34.81 Å / Num. obs: 46238 / % possible obs: 99.4 % / Redundancy: 5.5 % / CC1/2: 0.999 / Rmerge(I) obs: 0.062 / Rpim(I) all: 0.028 / Rrim(I) all: 0.069 / Net I/σ(I): 13.5 / Num. measured all: 256160 | ||||||||||||||||||
| Reflection shell |
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Starting model: pdbid 3C5K Resolution: 1.07→34.8 Å / Cor.coef. Fo:Fc: 0.979 / Cor.coef. Fo:Fc free: 0.978 / SU B: 0.931 / SU ML: 0.02 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.025 / ESU R Free: 0.024 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: Users of this crystal structure: verify our intepretion of the electron density. Amplitudes and unmerged intensities are included with this deposition. Diffraction images will be deposited ...Details: Users of this crystal structure: verify our intepretion of the electron density. Amplitudes and unmerged intensities are included with this deposition. Diffraction images will be deposited in a public repository. Geometry restraints for the fragment candidate were prepared with GRADE. Ambiguous difference density suggests some form of covalent modification of Cys1147.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 193.07 Å2 / Biso mean: 14.755 Å2 / Biso min: 7.96 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.07→34.8 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.07→1.098 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller










 PDBj
PDBj









