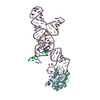
Entry Database : PDB / ID : 4wp2Title Chaetomium Mex67 UBA domain Putative mRNA export protein Keywords / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Chaetomium thermophilum (fungus)Method / / Resolution : 1.7 Å Authors Aibara, S. / Stewart, M. Funding support Organization Grant number Country Medical Research Council (United Kingdom)
Journal : Acta Crystallogr.,Sect.F / Year : 2015Title : Structural characterization of the principal mRNA-export factor Mex67-Mtr2 from Chaetomium thermophilum.Authors : Aibara, S. / Valkov, E. / Lamers, M.H. / Dimitrova, L. / Hurt, E. / Stewart, M. History Deposition Oct 17, 2014 Deposition site / Processing site Revision 1.0 Jul 15, 2015 Provider / Type Revision 2.0 Sep 13, 2017 Group / Atomic model / Author supporting evidenceCategory atom_site / pdbx_audit_support ... atom_site / pdbx_audit_support / pdbx_validate_close_contact / pdbx_validate_symm_contact Item _atom_site.B_iso_or_equiv / _atom_site.Cartn_x ... _atom_site.B_iso_or_equiv / _atom_site.Cartn_x / _atom_site.Cartn_y / _atom_site.Cartn_z / _pdbx_audit_support.funding_organization / _pdbx_validate_close_contact.auth_seq_id_1 / _pdbx_validate_close_contact.auth_seq_id_2 / _pdbx_validate_symm_contact.auth_seq_id_1 / _pdbx_validate_symm_contact.auth_seq_id_2 Revision 3.0 Nov 13, 2024 Group Atomic model / Data collection ... Atomic model / Data collection / Database references / Structure summary Category atom_site / chem_comp_atom ... atom_site / chem_comp_atom / chem_comp_bond / database_2 / pdbx_entry_details / pdbx_modification_feature Item / _database_2.pdbx_DOI / _database_2.pdbx_database_accession
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Chaetomium thermophilum (fungus)
Chaetomium thermophilum (fungus) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 1.7 Å
SYNCHROTRON / Resolution: 1.7 Å  Authors
Authors United Kingdom, 1items
United Kingdom, 1items  Citation
Citation Journal: Acta Crystallogr.,Sect.F / Year: 2015
Journal: Acta Crystallogr.,Sect.F / Year: 2015 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 4wp2.cif.gz
4wp2.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb4wp2.ent.gz
pdb4wp2.ent.gz PDB format
PDB format 4wp2.json.gz
4wp2.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/wp/4wp2
https://data.pdbj.org/pub/pdb/validation_reports/wp/4wp2 ftp://data.pdbj.org/pub/pdb/validation_reports/wp/4wp2
ftp://data.pdbj.org/pub/pdb/validation_reports/wp/4wp2 Links
Links Assembly
Assembly
 Components
Components Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 / Gene: CTHT_0059630 / Production host:
Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 / Gene: CTHT_0059630 / Production host: 
 X-RAY DIFFRACTION
X-RAY DIFFRACTION Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  Diamond
Diamond  / Beamline: I04-1 / Wavelength: 0.92 Å
/ Beamline: I04-1 / Wavelength: 0.92 Å Processing
Processing Movie
Movie Controller
Controller

















 PDBj
PDBj







