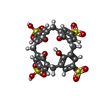
| Entry | Database: PDB / ID: 4pru
|
|---|
| Title | Crystal structure of dimethyllysine hen egg-white lysozyme in complex with sclx4 at 2.2 A resolution |
|---|
 Components Components | Lysozyme C |
|---|
 Keywords Keywords | HYDROLASE / HYDROLASE(O-GLYCOSYL) |
|---|
| Function / homology |  Function and homology information Function and homology information
Lactose synthesis / Antimicrobial peptides / Neutrophil degranulation / beta-N-acetylglucosaminidase activity / cell wall macromolecule catabolic process / lysozyme / lysozyme activity / killing of cells of another organism / defense response to Gram-negative bacterium / defense response to bacterium ...Lactose synthesis / Antimicrobial peptides / Neutrophil degranulation / beta-N-acetylglucosaminidase activity / cell wall macromolecule catabolic process / lysozyme / lysozyme activity / killing of cells of another organism / defense response to Gram-negative bacterium / defense response to bacterium / defense response to Gram-positive bacterium / endoplasmic reticulum / extracellular space / identical protein binding / cytoplasmSimilarity search - Function Lysozyme - #10 / Glycoside hydrolase, family 22, lysozyme / Glycoside hydrolase family 22 domain / Glycosyl hydrolases family 22 (GH22) domain signature. / Glycoside hydrolase, family 22 / C-type lysozyme/alpha-lactalbumin family / Glycosyl hydrolases family 22 (GH22) domain profile. / Alpha-lactalbumin / lysozyme C / Lysozyme / Lysozyme-like domain superfamily ...Lysozyme - #10 / Glycoside hydrolase, family 22, lysozyme / Glycoside hydrolase family 22 domain / Glycosyl hydrolases family 22 (GH22) domain signature. / Glycoside hydrolase, family 22 / C-type lysozyme/alpha-lactalbumin family / Glycosyl hydrolases family 22 (GH22) domain profile. / Alpha-lactalbumin / lysozyme C / Lysozyme / Lysozyme-like domain superfamily / Orthogonal Bundle / Mainly AlphaSimilarity search - Domain/homology |
|---|
| Biological species |   Gallus gallus (chicken) Gallus gallus (chicken) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å MOLECULAR REPLACEMENT / Resolution: 2.2 Å |
|---|
 Authors Authors | McGovern, R.E. / Lyons, J.A. / Crowley, P.B. |
|---|
 Citation Citation |  Journal: Chem Sci / Year: 2015 Journal: Chem Sci / Year: 2015
Title: Structural study of a small molecule receptor bound to dimethyllysine in lysozyme.
Authors: McGovern, R.E. / Snarr, B.D. / Lyons, J.A. / McFarlane, J. / Whiting, A.L. / Paci, I. / Hof, F. / Crowley, P.B. |
|---|
| History | | Deposition | Mar 6, 2014 | Deposition site: RCSB / Processing site: RCSB |
|---|
| Revision 1.0 | Nov 12, 2014 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Jan 21, 2015 | Group: Database references |
|---|
| Revision 1.2 | Sep 20, 2023 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_conn / struct_site
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_conn.pdbx_leaving_atom_flag / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å
MOLECULAR REPLACEMENT / Resolution: 2.2 Å  Authors
Authors Citation
Citation Journal: Chem Sci / Year: 2015
Journal: Chem Sci / Year: 2015 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 4pru.cif.gz
4pru.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb4pru.ent.gz
pdb4pru.ent.gz PDB format
PDB format 4pru.json.gz
4pru.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/pr/4pru
https://data.pdbj.org/pub/pdb/validation_reports/pr/4pru ftp://data.pdbj.org/pub/pdb/validation_reports/pr/4pru
ftp://data.pdbj.org/pub/pdb/validation_reports/pr/4pru

 Links
Links Assembly
Assembly


 Components
Components
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SLS
SLS  / Beamline: X10SA / Wavelength: 1.03319 Å
/ Beamline: X10SA / Wavelength: 1.03319 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj