[English] 日本語
 Yorodumi
Yorodumi- PDB-4nw2: Tandem chromodomains of human CHD1 in complex with Influenza viru... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4nw2 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Tandem chromodomains of human CHD1 in complex with Influenza virus NS1 C-terminal tail trimethylated at K229 | ||||||
 Components Components |
| ||||||
 Keywords Keywords | PEPTIDE BINDING PROTEIN/VIRAL PROTEIN / Structural Genomics Consortium / SGC / PEPTIDE BINDING PROTEIN-VIRAL PROTEIN complex | ||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host mRNA processing / symbiont-mediated suppression of host PKR/eIFalpha signaling / : / signaling receptor inhibitor activity / nucleosome organization / histone H3K4me3 reader activity / host-mediated activation of viral transcription / ATP-dependent chromatin remodeler activity / nuclear chromosome / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity ...symbiont-mediated suppression of host mRNA processing / symbiont-mediated suppression of host PKR/eIFalpha signaling / : / signaling receptor inhibitor activity / nucleosome organization / histone H3K4me3 reader activity / host-mediated activation of viral transcription / ATP-dependent chromatin remodeler activity / nuclear chromosome / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / protein serine/threonine kinase inhibitor activity / helicase activity / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / double-stranded RNA binding / histone binding / Estrogen-dependent gene expression / host cell cytoplasm / symbiont-mediated suppression of host innate immune response / chromatin remodeling / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / signaling receptor binding / symbiont-mediated suppression of host gene expression / chromatin binding / chromatin / host cell nucleus / ATP hydrolysis activity / DNA binding / RNA binding / nucleoplasm / ATP binding / identical protein binding / nucleus / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human)  Influenza A virus Influenza A virus | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.9 Å molecular replacement / Resolution: 1.9 Å | ||||||
 Authors Authors | Qin, S. / Tempel, W. / Xu, C. / El Bakkouri, M. / Bountra, C. / Arrowsmith, C.H. / Edwards, A.M. / Min, J. / Structural Genomics Consortium (SGC) | ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2014 Journal: Nat Commun / Year: 2014Title: Structural basis for histone mimicry and hijacking of host proteins by influenza virus protein NS1. Authors: Qin, S. / Liu, Y. / Tempel, W. / Eram, M.S. / Bian, C. / Liu, K. / Senisterra, G. / Crombet, L. / Vedadi, M. / Min, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4nw2.cif.gz 4nw2.cif.gz | 169.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4nw2.ent.gz pdb4nw2.ent.gz | 132.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4nw2.json.gz 4nw2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/nw/4nw2 https://data.pdbj.org/pub/pdb/validation_reports/nw/4nw2 ftp://data.pdbj.org/pub/pdb/validation_reports/nw/4nw2 ftp://data.pdbj.org/pub/pdb/validation_reports/nw/4nw2 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4o42C  4o45C  4o46C  2b2wS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 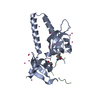
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 22942.408 Da / Num. of mol.: 2 / Fragment: UNP residues 268-443 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: CHD1 / Plasmid: pET28-MHL / Production host: Homo sapiens (human) / Gene: CHD1 / Plasmid: pET28-MHL / Production host:  #2: Protein/peptide | Mass: 1834.303 Da / Num. of mol.: 2 / Fragment: UNP residues 216-230 / Source method: obtained synthetically / Source: (synth.)   Influenza A virus / References: UniProt: T2F8K6, UniProt: P03495*PLUS Influenza A virus / References: UniProt: T2F8K6, UniProt: P03495*PLUS#3: Chemical | ChemComp-UNX / #4: Chemical | #5: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.49 Å3/Da / Density % sol: 50.54 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, sitting drop / pH: 8.5 Details: 10% PEG8000, 0.2M magnesium chloride, 0.1M Tris, pH 8.5, vapor diffusion, sitting drop, temperature 291K |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 19-ID / Wavelength: 0.97926 Å / Beamline: 19-ID / Wavelength: 0.97926 Å | ||||||||||||||||||
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: Jul 18, 2012 | ||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||
| Radiation wavelength | Wavelength: 0.97926 Å / Relative weight: 1 | ||||||||||||||||||
| Reflection | Resolution: 1.9→70.694 Å / Num. obs: 39818 / % possible obs: 100 % / Redundancy: 3.8 % / Biso Wilson estimate: 28.8 Å2 / Rmerge(I) obs: 0.075 / Net I/σ(I): 14.5 | ||||||||||||||||||
| Reflection shell | Rmerge(I) obs: 0.012 / Diffraction-ID: 1
|
-Phasing
| Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: pdb entry 2b2w Resolution: 1.9→42.968 Å / Occupancy max: 1 / Occupancy min: 0.5 / FOM work R set: 0.7981 / SU ML: 0.26 / σ(F): 0.12 / Phase error: 26.09 / Stereochemistry target values: ML Details: REFMAC WAS USED DURING INTERMEDIATE REFINEMENT STAGES. COOT WAS USED FOR INTERACTIVE MODEL BUILDING. MODEL GEOMETRY WAS ASSESSED ON THE MOLPROBITY SERVER. ELECTRON DENSITY FOR THE BACKBONE ...Details: REFMAC WAS USED DURING INTERMEDIATE REFINEMENT STAGES. COOT WAS USED FOR INTERACTIVE MODEL BUILDING. MODEL GEOMETRY WAS ASSESSED ON THE MOLPROBITY SERVER. ELECTRON DENSITY FOR THE BACKBONE OF THE PEPTIDE LIGAND IS NOT CONTINUOUS. IT IS THEREFORE POSSIBLE THAT THE PEPTIDE'S AMINO-TERMINUS IS MODELED IN A DIFFERENT ASYMMETRIC UNIT RELATIVE TO THE CARBOXY-TERMINUS. WEAK ELECTRON DENSITY NEAR P417 OF EITHER CHD1 CHAIN FAINTLY RESEMBLES A PEPTIDE.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 106.87 Å2 / Biso mean: 39.8335 Å2 / Biso min: 16.24 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.9→42.968 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 30
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj




