[English] 日本語
 Yorodumi
Yorodumi- PDB-4ntj: Structure of the human P2Y12 receptor in complex with an antithro... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4ntj | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of the human P2Y12 receptor in complex with an antithrombotic drug | ||||||
 Components Components | P2Y purinoceptor 12,Soluble cytochrome b562,P2Y purinoceptor 12 | ||||||
 Keywords Keywords | MEMBRANE PROTEIN / human P2Y12 receptor / GPCR network / lipidic cubic phase / antithrombotic drug / GPCR / PSI-Biology / Structural Genomics / Signaling Protein / membrane | ||||||
| Function / homology |  Function and homology information Function and homology informationvisual system development / positive regulation of integrin activation by cell surface receptor linked signal transduction / regulation of microglial cell migration / G protein-coupled ADP receptor activity / cerebral cortex radial glia-guided migration / cell body membrane / P2Y receptors / G protein-coupled purinergic nucleotide receptor activity / positive regulation of microglial cell migration / positive regulation of monoatomic ion transport ...visual system development / positive regulation of integrin activation by cell surface receptor linked signal transduction / regulation of microglial cell migration / G protein-coupled ADP receptor activity / cerebral cortex radial glia-guided migration / cell body membrane / P2Y receptors / G protein-coupled purinergic nucleotide receptor activity / positive regulation of microglial cell migration / positive regulation of monoatomic ion transport / G protein-coupled adenosine receptor activity / hemostasis / cell projection membrane / substrate-dependent cell migration, cell extension / regulation of chemotaxis / positive regulation of chemotaxis / cell projection organization / positive regulation of cell adhesion mediated by integrin / positive regulation of ruffle assembly / lamellipodium assembly / cellular response to ATP / response to axon injury / monoatomic ion transport / guanyl-nucleotide exchange factor activity / establishment of localization in cell / calcium-mediated signaling / electron transport chain / platelet activation / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / platelet aggregation / ADP signalling through P2Y purinoceptor 12 / G alpha (i) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / electron transfer activity / periplasmic space / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / iron ion binding / G protein-coupled receptor signaling pathway / heme binding / cell surface / membrane / plasma membrane Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.62 Å MOLECULAR REPLACEMENT / Resolution: 2.62 Å | ||||||
 Authors Authors | Zhang, K. / Zhang, J. / Gao, Z.-G. / Zhang, D. / Zhu, L. / Han, G.W. / Moss, S.M. / Paoletta, S. / Kiselev, E. / Lu, W. ...Zhang, K. / Zhang, J. / Gao, Z.-G. / Zhang, D. / Zhu, L. / Han, G.W. / Moss, S.M. / Paoletta, S. / Kiselev, E. / Lu, W. / Fenalti, G. / Zhang, W. / Muller, C.E. / Yang, H. / Jiang, H. / Cherezov, V. / Katritch, V. / Jacobson, K.A. / Stevens, R.C. / Wu, B. / Zhao, Q. / GPCR Network (GPCR) | ||||||
 Citation Citation |  Journal: Nature / Year: 2014 Journal: Nature / Year: 2014Title: Structure of the human P2Y12 receptor in complex with an antithrombotic drug Authors: Zhang, K. / Zhang, J. / Gao, Z.-G. / Zhang, D. / Zhu, L. / Han, G.W. / Moss, S.M. / Paoletta, S. / Kiselev, E. / Lu, W. / Fenalti, G. / Zhang, W. / Muller, C.E. / Yang, H. / Jiang, H. / ...Authors: Zhang, K. / Zhang, J. / Gao, Z.-G. / Zhang, D. / Zhu, L. / Han, G.W. / Moss, S.M. / Paoletta, S. / Kiselev, E. / Lu, W. / Fenalti, G. / Zhang, W. / Muller, C.E. / Yang, H. / Jiang, H. / Cherezov, V. / Katritch, V. / Jacobson, K.A. / Stevens, R.C. / Wu, B. / Zhao, Q. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4ntj.cif.gz 4ntj.cif.gz | 170.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4ntj.ent.gz pdb4ntj.ent.gz | 133.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4ntj.json.gz 4ntj.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/nt/4ntj https://data.pdbj.org/pub/pdb/validation_reports/nt/4ntj ftp://data.pdbj.org/pub/pdb/validation_reports/nt/4ntj ftp://data.pdbj.org/pub/pdb/validation_reports/nt/4ntj | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1m6tS  3vw7S S: Starting model for refinement |
|---|---|
| Similar structure data | |
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 53248.879 Da / Num. of mol.: 1 / Mutation: D294N, M1007W, H1102I, R1106L Source method: isolated from a genetically manipulated source Details: Chimera protein of N-terminal residues 2-223 from P2Y12R (P2Y12_HUMAN), Soluble cytochrome b562 (C562_ECOLX), and C-terminal residues 224-342 from P2Y12R (P2Y12_HUMAN). Source: (gene. exp.)  Homo sapiens (human), (gene. exp.) Homo sapiens (human), (gene. exp.)  Gene: P2RY12, HORK3, cybC / Plasmid: pFASTBAC1 / Production host:  | ||||||
|---|---|---|---|---|---|---|---|
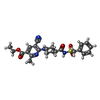
| #2: Chemical | ChemComp-AZJ / | ||||||
| #3: Chemical | | #4: Chemical | #5: Water | ChemComp-HOH / | Has protein modification | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 15 X-RAY DIFFRACTION / Number of used crystals: 15 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.23 Å3/Da / Density % sol: 61.91 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: lipidic cubic phase Details: 0.05-0.15M ammonium formate, 0.1M sodium cacodylate, pH 6.0-6.5, 25-35% PEG 400, 200M AZD1283, Lipidic Cubic Phase, temperature 293K PH range: 6.0-6.5 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 23-ID-D / Wavelength: 1.033 Å / Beamline: 23-ID-D / Wavelength: 1.033 Å |
| Detector | Type: MARMOSAIC 300 mm CCD / Detector: CCD / Date: Mar 23, 2013 / Details: mirror |
| Radiation | Monochromator: double crystal / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.033 Å / Relative weight: 1 |
| Reflection | Resolution: 2.6→50 Å / Num. obs: 19116 / % possible obs: 94.1 % / Redundancy: 3.8 % / Biso Wilson estimate: 84.46 Å2 / Rmerge(I) obs: 0.103 / Net I/σ(I): 14.4 |
| Reflection shell | Resolution: 2.6→2.69 Å / Redundancy: 3.1 % / Rmerge(I) obs: 0.958 / Mean I/σ(I) obs: 1.2 / Num. unique all: 1608 / % possible all: 79.5 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 3VW7 (PAR1), PDB ENTRY 1M6T (BRIL) Resolution: 2.62→26.44 Å / Cor.coef. Fo:Fc: 0.9456 / Cor.coef. Fo:Fc free: 0.9344 / Occupancy max: 1 / Occupancy min: 1 / SU R Cruickshank DPI: 0.385 / Cross valid method: THROUGHOUT / σ(F): 0 Details: THERE ARE SOME UNKNOWN DENSITIES. LOCATED NEAR THE SIDE CHAIN OF TYR 32. THEY HAVE NOT BEEN MODELLED.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 225.79 Å2 / Biso mean: 106.1 Å2 / Biso min: 64.32 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.549 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.62→26.44 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.62→2.76 Å / Total num. of bins used: 10
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj