[English] 日本語
 Yorodumi
Yorodumi- PDB-4nla: Structure of the central NEAT domain, N2, of the listerial Hbp2 p... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4nla | ||||||
|---|---|---|---|---|---|---|---|
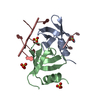
| Title | Structure of the central NEAT domain, N2, of the listerial Hbp2 protein, apo form | ||||||
 Components Components | Iron-regulated surface determinant protein A | ||||||
 Keywords Keywords | PROTEIN BINDING / Heme / NEAT domain / listeria / hemoglobin / N2 / Hbp / Hbp2 / HEME-BINDING PROTEIN / HEME BINDING | ||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to iron ion / iron ion transport / cell surface / extracellular region / metal ion binding / membrane Similarity search - Function | ||||||
| Biological species |  Listeria monocytogenes (bacteria) Listeria monocytogenes (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MAD / Resolution: 2.7 Å MAD / Resolution: 2.7 Å | ||||||
 Authors Authors | Malmirchegini, G.R. / Sawaya, M.R. / Clubb, R.T. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2014 Journal: J.Biol.Chem. / Year: 2014Title: Novel Mechanism of Hemin Capture by Hbp2, the Hemoglobin-binding Hemophore from Listeria monocytogenes. Authors: Malmirchegini, G.R. / Sjodt, M. / Shnitkind, S. / Sawaya, M.R. / Rosinski, J. / Newton, S.M. / Klebba, P.E. / Clubb, R.T. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4nla.cif.gz 4nla.cif.gz | 65.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4nla.ent.gz pdb4nla.ent.gz | 47.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4nla.json.gz 4nla.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  4nla_validation.pdf.gz 4nla_validation.pdf.gz | 437.1 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  4nla_full_validation.pdf.gz 4nla_full_validation.pdf.gz | 437.1 KB | Display | |
| Data in XML |  4nla_validation.xml.gz 4nla_validation.xml.gz | 6.8 KB | Display | |
| Data in CIF |  4nla_validation.cif.gz 4nla_validation.cif.gz | 8.1 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/nl/4nla https://data.pdbj.org/pub/pdb/validation_reports/nl/4nla ftp://data.pdbj.org/pub/pdb/validation_reports/nl/4nla ftp://data.pdbj.org/pub/pdb/validation_reports/nl/4nla | HTTPS FTP |
-Related structure data
| Related structure data |  4mypC  3sz6S C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
| ||||||||
| Details | The authors state that the biological unit is not known |
- Components
Components
| #1: Protein | Mass: 13545.126 Da / Num. of mol.: 1 / Fragment: unp residues 183-303 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Listeria monocytogenes (bacteria) / Strain: EGD-e / Gene: BN418_2635, lmo2185, M639_06195 / Plasmid: pHIS-SUMO / Production host: Listeria monocytogenes (bacteria) / Strain: EGD-e / Gene: BN418_2635, lmo2185, M639_06195 / Plasmid: pHIS-SUMO / Production host:  |
|---|---|
| #2: Chemical | ChemComp-SO4 / |
| #3: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.19 Å3/Da / Density % sol: 43.85 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: evaporation / pH: 5.4 Details: 100 mM sodium citrate tribasic dihydrate, 200 mM potassium sodium tartrate tetrahydrate, 2.0 M ammonium sulfate, pH 5.4, vapor diffusion, temperature 298K, EVAPORATION |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 24-ID-C / Wavelength: 0.97950, 0.9686, 1.5418 / Beamline: 24-ID-C / Wavelength: 0.97950, 0.9686, 1.5418 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Jun 6, 2013 Details: MIRRORS. BENT CYLINDERS, STRIPES OF PT, RH AND CLEAR. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Monochromator: SI 111 / Protocol: MAD / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Number: 126252 / Rmerge(I) obs: 0.057 / D res high: 2.69 Å / Num. obs: 6256 / % possible obs: 100 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Diffraction reflection shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.7→63.14 Å / Num. obs: 3370 / % possible obs: 100 % / Observed criterion σ(I): -3 / Redundancy: 21.4 % / Biso Wilson estimate: 109.36 Å2 / Rmerge(I) obs: 0.052 / Net I/σ(I): 35.3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Resolution: 2.7→2.85 Å / Redundancy: 22.8 % / Rmerge(I) obs: 0.471 / Mean I/σ(I) obs: 6.3 / % possible all: 100 |
-Phasing
| Phasing | Method:  MAD MAD | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phasing MAD | D res high: 2.6 Å / D res low: 19.85 Å / FOM : 0.432 / FOM acentric: 0.453 / FOM centric: 0.279 / Reflection: 3695 / Reflection acentric: 3245 / Reflection centric: 450 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Phasing MAD set | Highest resolution: 2.6 Å / Lowest resolution: 19.85 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Phasing MAD set shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Phasing MAD set site |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Phasing MAD shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Phasing dm | Method: Solvent flattening and Histogram matching / Reflection: 3695 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Phasing dm shell |
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MAD MADStarting model: PDB ENTRY 3SZ6 Resolution: 2.7→63.14 Å / Cor.coef. Fo:Fc: 0.939 / Cor.coef. Fo:Fc free: 0.926 / Occupancy max: 1 / Occupancy min: 1 / SU R Cruickshank DPI: 0.457 / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 120.65 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.968 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.7→63.14 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.7→3.02 Å / Total num. of bins used: 5
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: 16.3047 Å / Origin y: 37.9763 Å / Origin z: 28.8459 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group | Selection details: { A|* } |
 Movie
Movie Controller
Controller











 PDBj
PDBj



