[English] 日本語
 Yorodumi
Yorodumi- PDB-4mxf: X-ray structure of the adduct between bovine pancreatic ribonucle... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4mxf | ||||||
|---|---|---|---|---|---|---|---|
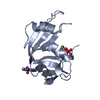
| Title | X-ray structure of the adduct between bovine pancreatic ribonuclease and Auoxo6, a dinuclear gold(III) complex with -dioxo bridges linking the two metal centers | ||||||
 Components Components | Ribonuclease pancreatic | ||||||
 Keywords Keywords | HYDROLASE / ribonuclease fold / RNA | ||||||
| Function / homology |  Function and homology information Function and homology informationpancreatic ribonuclease / ribonuclease A activity / RNA nuclease activity / nucleic acid binding / defense response to Gram-positive bacterium / hydrolase activity / extracellular region Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.25 Å MOLECULAR REPLACEMENT / Resolution: 2.25 Å | ||||||
 Authors Authors | Russo Krauss, I. / Vergara, A. / Merlino, A. | ||||||
 Citation Citation |  Journal: Metallomics / Year: 2014 Journal: Metallomics / Year: 2014Title: Interactions of gold-based drugs with proteins: crystal structure of the adduct formed between ribonuclease A and a cytotoxic gold(iii) compound. Authors: Messori, L. / Scaletti, F. / Massai, L. / Cinellu, M.A. / Russo Krauss, I. / di Martino, G. / Vergara, A. / Paduano, L. / Merlino, A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4mxf.cif.gz 4mxf.cif.gz | 61.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4mxf.ent.gz pdb4mxf.ent.gz | 45.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4mxf.json.gz 4mxf.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mx/4mxf https://data.pdbj.org/pub/pdb/validation_reports/mx/4mxf ftp://data.pdbj.org/pub/pdb/validation_reports/mx/4mxf ftp://data.pdbj.org/pub/pdb/validation_reports/mx/4mxf | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4l55S S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 13708.326 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #2: Chemical | ChemComp-AU / | #3: Water | ChemComp-HOH / | Has protein modification | Y | Nonpolymer details | CYTOTOXIC GOLD(III) COMPOUND IS NOT PRESENT IN THE STRUCTURE. THE STARTING METAL COMPLEX IS ...CYTOTOXIC GOLD(III) COMPOUND IS NOT PRESENT IN THE STRUCTURE. THE STARTING METAL COMPLEX IS COMPLETELY | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.96 Å3/Da / Density % sol: 37.3 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 5 Details: 20% PEG4000 and 20 mM sodium citrate buffer pH 5.0. Crystals appeared within 7 days. After two weeks, crystals were soaked in a solution of the gold-based drug dissolved in DMSO and in 50 mM ...Details: 20% PEG4000 and 20 mM sodium citrate buffer pH 5.0. Crystals appeared within 7 days. After two weeks, crystals were soaked in a solution of the gold-based drug dissolved in DMSO and in 50 mM sodium citrate buffer pH 5.0. At the final, concentrations of metallodrugs and protein were in at least 1:1 ratio., VAPOR DIFFUSION, HANGING DROP, temperature 298K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU / Wavelength: 1.54 Å ROTATING ANODE / Type: RIGAKU / Wavelength: 1.54 Å |
| Detector | Type: RIGAKU SATURN 944 / Detector: CCD / Date: Feb 5, 2013 |
| Radiation | Monochromator: GRAPHITE / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.54 Å / Relative weight: 1 |
| Reflection | Resolution: 2.25→23 Å / Num. all: 9860 / Num. obs: 9860 / % possible obs: 94.7 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 |
| Reflection shell | Resolution: 2.25→2.27 Å / % possible all: 94.9 |
- Processing
Processing
| Software | Name: REFMAC / Version: 5.7.0029 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entry 4L55 Resolution: 2.25→23 Å / Cor.coef. Fo:Fc: 0.951 / Cor.coef. Fo:Fc free: 0.9 / SU B: 9.368 / SU ML: 0.226 / Cross valid method: THROUGHOUT / ESU R: 0.541 / ESU R Free: 0.291 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 35.251 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.25→23 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.25→2.304 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller















 PDBj
PDBj






