[English] 日本語
 Yorodumi
Yorodumi- PDB-4mw1: Trypanosoma brucei methionyl-tRNA synthetase in complex with inhi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4mw1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3-chloro-5-methoxybenzyl)amino]propyl}-3-thiophen-3-ylurea (Chem 1444) | |||||||||
 Components Components | Methionyl-tRNA synthetase | |||||||||
 Keywords Keywords | LIGASE/LIGASE INHIBITOR / aminoacyl-tRNA synthetase / aaRS / MetRS / parasite / protein-inhibitor complex / Rossmann fold / translation / nucleotide binding / LIGASE-LIGASE INHIBITOR complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationmethionine-tRNA ligase / methionine-tRNA ligase activity / methionyl-tRNA aminoacylation / ciliary plasm / mitochondrion / ATP binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 2.494 Å molecular replacement / Resolution: 2.494 Å | |||||||||
 Authors Authors | Koh, C.Y. / Kim, J.E. / Wetzel, A.B. / de van der Schueren, W.J. / Shibata, S. / Liu, J. / Zhang, Z. / Fan, E. / Verlinde, C.L.M.J. / Hol, W.G.J. | |||||||||
 Citation Citation |  Journal: Plos Negl Trop Dis / Year: 2014 Journal: Plos Negl Trop Dis / Year: 2014Title: Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness. Authors: Koh, C.Y. / Kim, J.E. / Wetzel, A.B. / de van der Schueren, W.J. / Shibata, S. / Ranade, R.M. / Liu, J. / Zhang, Z. / Gillespie, J.R. / Buckner, F.S. / Verlinde, C.L. / Fan, E. / Hol, W.G. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4mw1.cif.gz 4mw1.cif.gz | 435.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4mw1.ent.gz pdb4mw1.ent.gz | 357.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4mw1.json.gz 4mw1.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mw/4mw1 https://data.pdbj.org/pub/pdb/validation_reports/mw/4mw1 ftp://data.pdbj.org/pub/pdb/validation_reports/mw/4mw1 ftp://data.pdbj.org/pub/pdb/validation_reports/mw/4mw1 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4mvwC  4mvxC  4mvyC  4mw0C  4mw2C  4mw4C  4mw5C  4mw6C  4mw7C  4mw9C  4mwbC  4mwcC  4mwdC  4mweC  4eg8S C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 61434.707 Da / Num. of mol.: 2 / Fragment: UNP residues 237-773 / Mutation: K456A, K457R, E458A Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|
-Non-polymers , 5 types, 443 molecules 


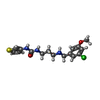





| #2: Chemical | ChemComp-GOL / #3: Chemical | ChemComp-DMS / #4: Chemical | ChemComp-MET / | #5: Chemical | ChemComp-44F / | #6: Water | ChemComp-HOH / | |
|---|
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.91 Å3/Da / Density % sol: 68.53 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, sitting drop Details: 2.0-2.3 M ammonium sulfate, 0.2 M sodium chloride, 0.1 M sodium cacodylate, pH 6.0-6.8, VAPOR DIFFUSION, SITTING DROP, temperature 298K PH range: 6.0-6.8 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 8.2.1 / Wavelength: 1 Å / Beamline: 8.2.1 / Wavelength: 1 Å |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Dec 3, 2010 |
| Diffraction measurement | Details: 1.00 degrees, 3.0 sec, detector distance 295.00 mm |
| Radiation | Monochromator: double crystal Si(111) / Protocol: SINGLE WAVELENGTH / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Av R equivalents: 0.16 / Number: 497084 |
| Reflection | Resolution: 2.494→50 Å / Num. obs: 67803 / % possible obs: 100 % / Observed criterion σ(F): 0 / Observed criterion σ(I): -3 / Redundancy: 7.3 % / Rmerge(I) obs: 0.16 / Rsym value: 0.16 / Χ2: 0.953 / Net I/σ(I): 5.4 |
| Reflection shell | Resolution: 2.494→2.54 Å / Redundancy: 7.4 % / Rmerge(I) obs: 0.852 / Mean I/σ(I) obs: 1.875 / Rsym value: 0.852 / % possible all: 100 |
| Cell measurement | Reflection used: 497084 |
-Phasing
| Phasing | Method:  molecular replacement molecular replacement | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phasing MR | Rfactor: 32.46 / Model details: Phaser MODE: MR_AUTO
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 4EG8 Resolution: 2.494→30 Å / Cor.coef. Fo:Fc: 0.941 / Cor.coef. Fo:Fc free: 0.924 / WRfactor Rfree: 0.197 / WRfactor Rwork: 0.171 / Occupancy max: 1 / Occupancy min: 0.3 / SU B: 12.661 / SU ML: 0.148 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.278 / ESU R Free: 0.212 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: BABINET MODEL WITH MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 132.03 Å2 / Biso mean: 35.617 Å2 / Biso min: 11.2 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.494→30 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 20
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller















 PDBj
PDBj




