[English] 日本語
 Yorodumi
Yorodumi- PDB-4mvn: Crystal structure of the staphylococcal serine protease SplA in c... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4mvn | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the staphylococcal serine protease SplA in complex with a specific phosphonate inhibitor | ||||||
 Components Components | Serine protease splA | ||||||
 Keywords Keywords | HYDROLASE/HYDROLASE INHIBITOR / CHYMOTRYPSIN-LIKE FOLD / SERINE ENDOPEPTIDASE / EXTRACELLULAR STAPHYLOCOCCAL PROTEASES / HYDROLASE-HYDROLASE INHIBITOR COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationHydrolases; Acting on peptide bonds (peptidases); Serine endopeptidases / serine-type endopeptidase activity / proteolysis / extracellular region Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.7 Å MOLECULAR REPLACEMENT / Resolution: 1.7 Å | ||||||
 Authors Authors | Zdzalik, M. / Burchacka, E. / Niemczyk, J.S. / Pustelny, K. / Popowicz, G.M. / Wladyka, B. / Dubin, A. / Potempa, J. / Sienczyk, M. / Dubin, G. / Oleksyszyn, J. | ||||||
 Citation Citation |  Journal: Protein Sci. / Year: 2014 Journal: Protein Sci. / Year: 2014Title: Development and binding characteristics of phosphonate inhibitors of SplA protease from Staphylococcus aureus. Authors: Burchacka, E. / Zdzalik, M. / Niemczyk, J.S. / Pustelny, K. / Popowicz, G. / Wladyka, B. / Dubin, A. / Potempa, J. / Sienczyk, M. / Dubin, G. / Oleksyszyn, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4mvn.cif.gz 4mvn.cif.gz | 182.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4mvn.ent.gz pdb4mvn.ent.gz | 146.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4mvn.json.gz 4mvn.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mv/4mvn https://data.pdbj.org/pub/pdb/validation_reports/mv/4mvn ftp://data.pdbj.org/pub/pdb/validation_reports/mv/4mvn ftp://data.pdbj.org/pub/pdb/validation_reports/mv/4mvn | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 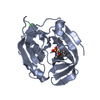 3ufaC  2w7sS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 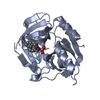
| ||||||||
| 2 | 
| ||||||||
| 3 | 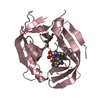
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 21885.482 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: Q2FXC2, Hydrolases; Acting on peptide bonds (peptidases); Serine endopeptidases #2: Chemical | ChemComp-I1S / [( #3: Water | ChemComp-HOH / | Has protein modification | Y | Nonpolymer details | THE INHIBITOR FULL CHEMICAL FORMULA CBZ-PHE(P)(OC6H5-4-SO2CH3)2 IS HYDROLYSED BY THE PROTEASE (THAT ...THE INHIBITOR FULL CHEMICAL FORMULA CBZ-PHE(P)(OC6H5-4-SO2CH3)2 IS HYDROLYSED | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.28 Å3/Da / Density % sol: 46.07 % |
|---|---|
| Crystal grow | Method: vapor diffusion, sitting drop / pH: 7 Details: 0.1M HEPES, 0.2M CALCIUM CHLORIDE, 25% PEG 4000, PH 7.0, VAPOR DIFFUSION, TEMPERATURE 297K, VAPOR DIFFUSION, SITTING DROP |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X10SA / Wavelength: 1 / Beamline: X10SA / Wavelength: 1 |
| Detector | Type: MAR CCD 165 mm / Detector: CCD / Date: Jul 14, 2008 |
| Radiation | Monochromator: SI(111) MONOCHROMATOR / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.7→25 Å / Num. obs: 85873 / % possible obs: 96.7 % |
| Reflection shell | Resolution: 1.7→1.74 Å |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 2W7S Resolution: 1.7→25 Å / Cor.coef. Fo:Fc: 0.963 / Cor.coef. Fo:Fc free: 0.943 / SU B: 1.912 / SU ML: 0.065 / Cross valid method: THROUGHOUT / ESU R: 0.105 / ESU R Free: 0.108 / Stereochemistry target values: MAXIMUM LIKELIHOOD
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 19.44 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.7→25 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj

