[English] 日本語
 Yorodumi
Yorodumi- PDB-4mfp: The crystal structure of acyltransferase in complex with decanoyl... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4mfp | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The crystal structure of acyltransferase in complex with decanoyl-CoA and Tei pseudoaglycone | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | Transferase/Antibiotic / GNAT / acyltransferase / acyl-CoA / Transferase-Antibiotic complex | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Actinoplanes teichomyceticus (bacteria) Actinoplanes teichomyceticus (bacteria) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.15 Å MOLECULAR REPLACEMENT / Resolution: 2.15 Å | |||||||||
 Authors Authors | Lyu, S.Y. / Liu, Y.C. / Chang, C.Y. / Huang, C.J. / Li, T.L. | |||||||||
 Citation Citation |  Journal: J.Am.Chem.Soc. / Year: 2014 Journal: J.Am.Chem.Soc. / Year: 2014Title: Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant enterococcus Authors: Lyu, S.Y. / Liu, Y.C. / Chang, C.Y. / Huang, C.J. / Chiu, Y.H. / Huang, C.M. / Hsu, N.S. / Lin, K.H. / Wu, C.J. / Tsai, M.D. / Li, T.L. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4mfp.cif.gz 4mfp.cif.gz | 97.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4mfp.ent.gz pdb4mfp.ent.gz | 71.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4mfp.json.gz 4mfp.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mf/4mfp https://data.pdbj.org/pub/pdb/validation_reports/mf/4mfp ftp://data.pdbj.org/pub/pdb/validation_reports/mf/4mfp ftp://data.pdbj.org/pub/pdb/validation_reports/mf/4mfp | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4mfjSC  4mfkC  4mflC  4mfqC  4mfzC  4q36C  4q38C  4mfs  4mft  4mfw  4mfx  4mfy  4mg0  4mg1 S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein / Protein/peptide , 2 types, 2 molecules AB
| #1: Protein | Mass: 39204.344 Da / Num. of mol.: 1 / Mutation: H196A Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Actinoplanes teichomyceticus (bacteria) Actinoplanes teichomyceticus (bacteria)Gene: tcp24 / Production host:  References: UniProt: Q70AY4, Transferases; Acyltransferases; Transferring groups other than aminoacyl groups |
|---|---|
| #2: Protein/peptide | |
-Sugars , 3 types, 3 molecules 

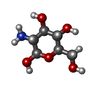


| #5: Sugar | ChemComp-NAG / |
|---|---|
| #6: Sugar | ChemComp-MAN / |
| #7: Sugar | ChemComp-GCS / |
-Non-polymers , 4 types, 258 molecules 

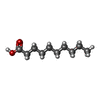




| #3: Chemical | ChemComp-COA / | ||||
|---|---|---|---|---|---|
| #4: Chemical | | #8: Chemical | ChemComp-DKA / | #9: Water | ChemComp-HOH / | |
-Details
| Nonpolymer details | ABOUT THE BOND DISTANCE BETWEEN N2 (GCS 503) AND C1 (DKA 504) IS HIGHER THAN CUTOFF DISTANCE, THE ...ABOUT THE BOND DISTANCE BETWEEN N2 (GCS 503) AND C1 (DKA 504) IS HIGHER THAN CUTOFF DISTANCE, THE DEPOSITORS |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.18 Å3/Da / Density % sol: 61.27 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 6.5 Details: 0.1mM MES, 0.2M ammonium sulphate, 30%(V/V) PEG5000 MME, pH 6.5, VAPOR DIFFUSION, HANGING DROP, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSRRC NSRRC  / Beamline: BL15A / Wavelength: 1 Å / Beamline: BL15A / Wavelength: 1 Å |
| Detector | Type: RAYONIX MX300HE / Detector: CCD / Date: Mar 19, 2013 |
| Radiation | Monochromator: LN2-Cooled, Fixed-Exit Double Crystal Monochromator Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.15→30 Å / Num. all: 26394 / Num. obs: 26394 / % possible obs: 99.9 % / Observed criterion σ(F): 2 / Observed criterion σ(I): 2 |
| Reflection shell | Resolution: 2.15→2.23 Å / % possible all: 100 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4MFJ Resolution: 2.15→30 Å / Cor.coef. Fo:Fc: 0.962 / Cor.coef. Fo:Fc free: 0.947 / SU B: 4.658 / SU ML: 0.121 / Cross valid method: THROUGHOUT / σ(F): 2 / ESU R: 0.185 / ESU R Free: 0.164 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN USED IF PRESENT IN THE INPUT
| |||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 40.945 Å2
| |||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.15→30 Å
| |||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.153→2.209 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller












 PDBj
PDBj