Entry Database : PDB / ID : 4k7dTitle Crystal Structure of Parkin C-terminal RING domains E3 ubiquitin-protein ligase parkin Keywords / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Rattus norvegicus (Norway rat)Method / / / Resolution : 2.8 Å Authors Sauve, V. / Trempe, J.-F. / Menade, M. / Gehring, K. Journal : Science / Year : 2013Title : Structure of parkin reveals mechanisms for ubiquitin ligase activation.Authors : Trempe, J.F. / Sauve, V. / Grenier, K. / Seirafi, M. / Tang, M.Y. / Menade, M. / Al-Abdul-Wahid, S. / Krett, J. / Wong, K. / Kozlov, G. / Nagar, B. / Fon, E.A. / Gehring, K. History Deposition Apr 17, 2013 Deposition site / Processing site Revision 1.0 May 15, 2013 Provider / Type Revision 1.1 May 22, 2013 Group Revision 1.2 Jul 10, 2013 Group Revision 1.3 Feb 28, 2024 Group / Database references / Derived calculationsCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_struct_conn_angle / struct_conn / struct_ref_seq_dif / struct_site Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_asym_id / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr2_auth_asym_id / _pdbx_struct_conn_angle.ptnr2_auth_seq_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_ref_seq_dif.details / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  SAD / Resolution: 2.8 Å
SAD / Resolution: 2.8 Å  Authors
Authors Citation
Citation Journal: Science / Year: 2013
Journal: Science / Year: 2013 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 4k7d.cif.gz
4k7d.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb4k7d.ent.gz
pdb4k7d.ent.gz PDB format
PDB format 4k7d.json.gz
4k7d.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/k7/4k7d
https://data.pdbj.org/pub/pdb/validation_reports/k7/4k7d ftp://data.pdbj.org/pub/pdb/validation_reports/k7/4k7d
ftp://data.pdbj.org/pub/pdb/validation_reports/k7/4k7d Links
Links Assembly
Assembly



 Components
Components

 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  CLSI
CLSI  / Beamline: 08ID-1 / Wavelength: 1.2824 Å
/ Beamline: 08ID-1 / Wavelength: 1.2824 Å SAD
SAD Processing
Processing SAD / Resolution: 2.8→53.31 Å / Occupancy max: 1 / Occupancy min: 0.5 / FOM work R set: 0.8301 / SU ML: 0.32 / σ(F): 1.36 / Phase error: 23.44 / Stereochemistry target values: ML
SAD / Resolution: 2.8→53.31 Å / Occupancy max: 1 / Occupancy min: 0.5 / FOM work R set: 0.8301 / SU ML: 0.32 / σ(F): 1.36 / Phase error: 23.44 / Stereochemistry target values: ML Movie
Movie Controller
Controller






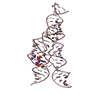

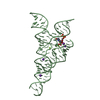





 PDBj
PDBj








