[English] 日本語
 Yorodumi
Yorodumi- PDB-4j7d: The 1.25A crystal structure of humanized Xenopus MDM2 with a nutl... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4j7d | ||||||
|---|---|---|---|---|---|---|---|
| Title | The 1.25A crystal structure of humanized Xenopus MDM2 with a nutlin fragment, RO5045331 | ||||||
 Components Components | E3 ubiquitin-protein ligase Mdm2 | ||||||
 Keywords Keywords | LIGASE/ANTAGONIST / PROTEIN-PROTEIN INTERACTION / E3 UBIQUITIN LIGASE / LIGASE-ANTAGONIST COMPLEX / P53 / IMIDAZOLINE / NUCLEUS | ||||||
| Function / homology |  Function and homology information Function and homology informationregulation of biological quality / positive regulation of mitotic cell cycle / RING-type E3 ubiquitin transferase / p53 binding / ubiquitin protein ligase activity / regulation of gene expression / ubiquitin-dependent protein catabolic process / protein ubiquitination / apoptotic process / negative regulation of apoptotic process ...regulation of biological quality / positive regulation of mitotic cell cycle / RING-type E3 ubiquitin transferase / p53 binding / ubiquitin protein ligase activity / regulation of gene expression / ubiquitin-dependent protein catabolic process / protein ubiquitination / apoptotic process / negative regulation of apoptotic process / nucleolus / zinc ion binding / nucleoplasm / identical protein binding / nucleus / cytoplasm Similarity search - Function | ||||||
| Biological species | |||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.25 Å MOLECULAR REPLACEMENT / Resolution: 1.25 Å | ||||||
 Authors Authors | Janson, C. / Lukacs, C. / Graves, B. | ||||||
 Citation Citation |  Journal: ACS Med Chem Lett / Year: 2013 Journal: ACS Med Chem Lett / Year: 2013Title: Deconstruction of a nutlin: dissecting the binding determinants of a potent protein-protein interaction inhibitor. Authors: Fry, D.C. / Wartchow, C. / Graves, B. / Janson, C. / Lukacs, C. / Kammlott, U. / Belunis, C. / Palme, S. / Klein, C. / Vu, B. #1:  Journal: To be Published / Year: 2013 Journal: To be Published / Year: 2013Title: MDM2 small molecule antagonist RG7112 activates P53 signaling and regresses human tumors in preclinical cancer models Authors: Tovar, C. / Graves, B. / Packman, K. / Filipovic, Z. / Higgins, B. / Xia, M. / Tardell, C. / Garrido, R. / Lee, E. / Kolinsky, K. / To, K.-H. / Linn, M. / Podlaski, F. / Wovkulich, P. / Vu, B. / Vassilev, L.T. #2:  Journal: To be Published Journal: To be PublishedTitle: Discovery of RG7112: a small-molecule mdm2 antagonist in clinical development Authors: Vu, B. / Wovkulich, P. / Pizzolato, G. / Lovey, A. / Ding, Q. / Jiang, N. / Liu, J.-J. / Zhao, C. / Glenn, K. / Wen, Y. / Tovar, C. / Thompson, T. / Packman, K. / Vassilev, L. / Graves, B. #3:  Journal: Science / Year: 2004 Journal: Science / Year: 2004Title: In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Authors: Vassilev, L.T. / Vu, B.T. / Graves, B. / Carvajal, D. / Podlaski, F. / Filipovic, Z. / Kong, N. / Kammlott, U. / Lukacs, C. / Klein, C. / Fotouhi, N. / Liu, E.A. | ||||||
| History |
|
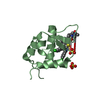
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4j7d.cif.gz 4j7d.cif.gz | 35.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4j7d.ent.gz pdb4j7d.ent.gz | 22.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4j7d.json.gz 4j7d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/j7/4j7d https://data.pdbj.org/pub/pdb/validation_reports/j7/4j7d ftp://data.pdbj.org/pub/pdb/validation_reports/j7/4j7d ftp://data.pdbj.org/pub/pdb/validation_reports/j7/4j7d | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Details | N-TERMINAL FRAGMENT EXISTS AS A MONOMER BUT FULL-LENGTH PROTEIN WOULD BE A DIMER |
- Components
Components
| #1: Protein | Mass: 9962.615 Da / Num. of mol.: 1 / Fragment: N-terminal domain (UNP residues 21-105) / Mutation: I50L, P92H, L95I Source method: isolated from a genetically manipulated source Source: (gene. exp.)  References: UniProt: P56273, Ligases; Forming carbon-nitrogen bonds; Acid-amino-acid ligases (peptide synthases) | ||
|---|---|---|---|
| #2: Chemical | ChemComp-I31 / ( | ||
| #3: Chemical | | #4: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.09 Å3/Da / Density % sol: 41.18 % |
|---|---|
| Crystal grow | Temperature: 278 K / Method: vapor diffusion, hanging drop / pH: 6.5 Details: 40-50% SATURATED AMMONIUM SULFATE, 0.1M MES, PH 6.5, 5% PEG200, VAPOR DIFFUSION, HANGING DROP, temperature 278K |
-Data collection
| Diffraction | Mean temperature: 80 K | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X10SA / Wavelength: 1 Å / Beamline: X10SA / Wavelength: 1 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: PSI PILATUS 6M / Detector: PIXEL / Date: Sep 22, 2012 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.17→40 Å / Num. all: 28271 / Num. obs: 27483 / % possible obs: 98.2 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 6.1 % / Biso Wilson estimate: 10.8 Å2 / Rmerge(I) obs: 0.044 / Net I/σ(I): 9.9 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 1.25→34.51 Å / Rfactor Rfree error: 0.007 / Data cutoff high absF: 902998.51 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber MOLECULAR REPLACEMENT / Resolution: 1.25→34.51 Å / Rfactor Rfree error: 0.007 / Data cutoff high absF: 902998.51 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 34.4755 Å2 / ksol: 0.340739 e/Å3 | ||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 14.9 Å2
| ||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.25→34.51 Å
| ||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | NCS model details: NONE | ||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.25→1.33 Å / Rfactor Rfree error: 0.018 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||
| Xplor file |
|
 Movie
Movie Controller
Controller















 PDBj
PDBj