Entry Database : PDB / ID : 4bewTitle SERCA bound to phosphate analogue SARCOPLASMIC/ENDOPLASMIC RETICULUM CALCIUM ATPASE 1 Keywords / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species ORYCTOLAGUS CUNICULUS (rabbit)Method / / / Resolution : 2.5 Å Authors Drachmann, N.D. / Mattle, D. / Laursen, M. / Bublitz, M. / Olesen, C. / Moeller, J.V. / Nissen, P. / Morth, J.P. Journal : To be Published Title : Serca Bound to Phosphate AnalogueAuthors : Mattle, D. / Drachmann, N.D. / Liu, X.Y. / Gourdon, P. / Pedersen, B.P. / Morth, P. / Wang, J. / Nissen, P. History Deposition Mar 12, 2013 Deposition site / Processing site Revision 1.0 Feb 19, 2014 Provider / Type Revision 1.1 Jan 30, 2019 Group / Experimental preparation / OtherCategory / pdbx_database_proc / pdbx_database_statusItem / _pdbx_database_status.recvd_author_approvalRevision 1.2 Feb 6, 2019 Group / Experimental preparation / Category / Item Revision 1.3 Dec 20, 2023 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Other / Refinement description Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_database_status / pdbx_initial_refinement_model / pdbx_struct_conn_angle / struct_conn / struct_site Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_database_status.status_code_sf / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr2_auth_comp_id / _pdbx_struct_conn_angle.ptnr2_auth_seq_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr2_label_atom_id / _pdbx_struct_conn_angle.ptnr2_label_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id Revision 1.4 Nov 20, 2024 Group / Category / pdbx_modification_feature
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.5 Å
MOLECULAR REPLACEMENT / Resolution: 2.5 Å  Authors
Authors Citation
Citation Journal: To be Published
Journal: To be Published Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 4bew.cif.gz
4bew.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb4bew.ent.gz
pdb4bew.ent.gz PDB format
PDB format 4bew.json.gz
4bew.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/be/4bew
https://data.pdbj.org/pub/pdb/validation_reports/be/4bew ftp://data.pdbj.org/pub/pdb/validation_reports/be/4bew
ftp://data.pdbj.org/pub/pdb/validation_reports/be/4bew
 Links
Links Assembly
Assembly


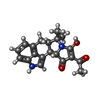
 Components
Components












 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SLS
SLS  / Beamline: X06SA / Wavelength: 1
/ Beamline: X06SA / Wavelength: 1  Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller






















 PDBj
PDBj











