[English] 日本語
 Yorodumi
Yorodumi- PDB-4b3v: Crystal structure of the Rubella virus glycoprotein E1 in its pos... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4b3v | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the Rubella virus glycoprotein E1 in its post-fusion form crystallized in presence of 20mM of Calcium Acetate | ||||||
 Components Components | E1 ENVELOPE GLYCOPROTEIN | ||||||
 Keywords Keywords | VIRAL PROTEIN / ENVELOPE GLYCOPROTEIN / MEMBRANE FUSION | ||||||
| Function / homology |  Function and homology information Function and homology informationT=4 icosahedral viral capsid / host cell Golgi membrane / host cell mitochondrion / viral nucleocapsid / clathrin-dependent endocytosis of virus by host cell / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / virion membrane / RNA binding ...T=4 icosahedral viral capsid / host cell Golgi membrane / host cell mitochondrion / viral nucleocapsid / clathrin-dependent endocytosis of virus by host cell / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / virion membrane / RNA binding / metal ion binding / membrane Similarity search - Function | ||||||
| Biological species |  RUBELLA VIRUS RUBELLA VIRUS | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.98 Å MOLECULAR REPLACEMENT / Resolution: 1.98 Å | ||||||
 Authors Authors | Vaney, M.C. / DuBois, R.M. / Tortorici, M.A. / Rey, F.A. | ||||||
 Citation Citation |  Journal: Nature / Year: 2013 Journal: Nature / Year: 2013Title: Functional and Evolutionary Insight from the Crystal Structure of Rubella Virus Protein E1. Authors: Dubois, R.M. / Vaney, M.C. / Tortorici, M.A. / Kurdi, R.A. / Barba-Spaeth, G. / Krey, T. / Rey, F.A. #1:  Journal: Cell(Cambridge,Mass.) / Year: 2001 Journal: Cell(Cambridge,Mass.) / Year: 2001Title: The Fusion Glycoprotein Shell of Semliki Forest Virus: An Icosahedral Assembly Primed for Fusogenic Activation at Endosomal Ph. Authors: Lescar, J. / Roussel, A. / Wien, M.W. / Navaza, J. / Fuller, S.D. / Wengler, G. / Wengler, G. / Rey, F.A. #2:  Journal: Nature / Year: 2004 Journal: Nature / Year: 2004Title: Conformational Change and Protein-Protein Interactions of the Fusion Protein of Semliki Forest Virus. Authors: Gibbons, D.L. / Vaney, M. / Roussel, A. / Vigouroux, A. / Reilly, B. / Lepault, J. / Kielian, M. / Rey, F.A. #3:  Journal: Structure / Year: 2006 Journal: Structure / Year: 2006Title: Structure and Interactions at the Viral Surface of the Envelope Protein E1 of Semliki Forest Virus. Authors: Roussel, A. / Lescar, J. / Vaney, M. / Wengler, G. / Wengler, G. / Rey, F.A. #4:  Journal: Embo J. / Year: 2004 Journal: Embo J. / Year: 2004Title: Structure of a Flavivirus Envelope Glycoprotein in its Low-Ph-Induced Membrane Fusion Conformation. Authors: Bressanelli, S. / Stiasny, K. / Allison, S.L. / Stura, E.A. / Duquerroy, S. / Lescar, J. / Heinz, F.X. / Rey, F.A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4b3v.cif.gz 4b3v.cif.gz | 526.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4b3v.ent.gz pdb4b3v.ent.gz | 431 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4b3v.json.gz 4b3v.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/b3/4b3v https://data.pdbj.org/pub/pdb/validation_reports/b3/4b3v ftp://data.pdbj.org/pub/pdb/validation_reports/b3/4b3v ftp://data.pdbj.org/pub/pdb/validation_reports/b3/4b3v | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4adgC  4adiSC  4adjC C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 3 molecules ABC
| #1: Protein | Mass: 50573.352 Da / Num. of mol.: 3 / Fragment: ECTODOMAIN, RESIDUES 583-1018 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  RUBELLA VIRUS / Strain: M33 / Plasmid: PMT MODIFIED INVITROGEN / Cell line (production host): SCHNEIDER 2 / Production host: RUBELLA VIRUS / Strain: M33 / Plasmid: PMT MODIFIED INVITROGEN / Cell line (production host): SCHNEIDER 2 / Production host:  |
|---|
-Sugars , 2 types, 6 molecules 
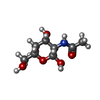

| #2: Sugar | ChemComp-NAG / #3: Sugar | |
|---|
-Non-polymers , 7 types, 1162 molecules 












| #4: Chemical | ChemComp-GOL / #5: Chemical | ChemComp-ACT / #6: Chemical | ChemComp-PG4 / | #7: Chemical | ChemComp-PEG / #8: Chemical | ChemComp-NA / | #9: Chemical | #10: Water | ChemComp-HOH / | |
|---|
-Details
| Has protein modification | Y |
|---|---|
| Nonpolymer details | CALCIUM ION (CA): THE CALCIUM IONS ARE BONDED TO RESIDUES AT THE FUSION LOOPS OF THREE PROTOMERS. ...CALCIUM ION (CA): THE CALCIUM IONS ARE BONDED TO RESIDUES AT THE FUSION LOOPS OF THREE PROTOMERS. SODIUM ION (NA): THE SODIUM ION IS BONDED TO RESIDUES AT THE FUSION LOOPS OF ONE PROTOMER. GLYCEROL (GOL): E1 PROTEIN WAS CRYSTALLIZ |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3 Å3/Da / Density % sol: 59 % / Description: NONE |
|---|---|
| Crystal grow | Details: 1-6% PEG 4K, 100 MM NAHEPES PH 8, 25% GLYCEROL, 20 MM CA(OAC)2 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SOLEIL SOLEIL  / Beamline: PROXIMA 1 / Wavelength: 1 / Beamline: PROXIMA 1 / Wavelength: 1 |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Jul 2, 2010 / Details: DYNAMICALLY BENDABLE |
| Radiation | Monochromator: DOUBLE-CRYSTAL FIXED-EXIT / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.98→43.72 Å / Num. obs: 135009 / % possible obs: 97.1 % / Observed criterion σ(I): 2 / Redundancy: 3.4 % / Biso Wilson estimate: 29.81 Å2 / Rmerge(I) obs: 0.08 / Net I/σ(I): 9.1 |
| Reflection shell | Resolution: 1.98→2.08 Å / Redundancy: 2.6 % / Rmerge(I) obs: 0.5 / Mean I/σ(I) obs: 2 / % possible all: 81.5 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 4ADI Resolution: 1.98→28.99 Å / Cor.coef. Fo:Fc: 0.9503 / Cor.coef. Fo:Fc free: 0.946 / SU R Cruickshank DPI: 0.13 / Cross valid method: THROUGHOUT / σ(F): 0 / SU R Blow DPI: 0.137 / SU Rfree Blow DPI: 0.117 / SU Rfree Cruickshank DPI: 0.114 Details: REFINEMENT NOTE 1, IDEAL-DIST CONTACT TERM CONTACT SETUP. RESIDUE TYPES WITHOUT CCP4 ATOM TYPE IN LIBRARY=NA CA. NUMBER OF ATOMS WITH PROPER CCP4 ATOM TYPE=11228. NUMBER WITH APPROX DEFAULT ...Details: REFINEMENT NOTE 1, IDEAL-DIST CONTACT TERM CONTACT SETUP. RESIDUE TYPES WITHOUT CCP4 ATOM TYPE IN LIBRARY=NA CA. NUMBER OF ATOMS WITH PROPER CCP4 ATOM TYPE=11228. NUMBER WITH APPROX DEFAULT CCP4 ATOM TYPE=0. NUMBER TREATED BY BAD NON-BONDED CONTACTS=4.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 33.11 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.242 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.98→28.99 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.98→2.03 Å / Total num. of bins used: 20
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller










 PDBj
PDBj







