[English] 日本語
 Yorodumi
Yorodumi- PDB-480d: CRYSTAL STRUCTURE OF THE SARCIN/RICIN DOMAIN FROM E. COLI 23 S RRNA -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 480d | ||||||
|---|---|---|---|---|---|---|---|
| Title | CRYSTAL STRUCTURE OF THE SARCIN/RICIN DOMAIN FROM E. COLI 23 S RRNA | ||||||
 Components Components | SARCIN/RICIN DOMAIN FROM 23 S RRNA | ||||||
 Keywords Keywords | RNA / SARCIN/RICIN LOOP OR DOMAIN / RNA RECOGNITION / RIBOSOMES / ELONGATION FACTORS | ||||||
| Function / homology | RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.5 Å MOLECULAR REPLACEMENT / Resolution: 1.5 Å | ||||||
 Authors Authors | Correll, C.C. / Wool, I.G. / Munishkin, A. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1999 Journal: J.Mol.Biol. / Year: 1999Title: The two faces of the Escherichia coli 23 S rRNA sarcin/ricin domain: the structure at 1.11 A resolution. Authors: Correll, C.C. / Wool, I.G. / Munishkin, A. #1:  Journal: Proc.Natl.Acad.Sci.USA / Year: 1998 Journal: Proc.Natl.Acad.Sci.USA / Year: 1998Title: Crystal structure of the ribosomal RNA domain essential for binding elongation factors Authors: Correll, C.C. / Munishkin, A. / Chan, Y.-L. / Ren, Z. / Wool, I.G. #2:  Journal: Biophys.J. / Year: 1999 Journal: Biophys.J. / Year: 1999Title: Comparison of the crystal and solution structures of two RNA oligonucleotides Authors: Rife, J.P. / Stallings, S.G. / Correll, C.C. / Dallas, A. / Steitz, T.A. / Moore, P.B. #3:  Journal: J.Mol.Biol. / Year: 1995 Journal: J.Mol.Biol. / Year: 1995Title: The sarcin/ricin loop, a modular RNA Authors: Szewczak, A.A. / Moore, P.B. #4:  Journal: Proc.Natl.Acad.Sci.USA / Year: 1993 Journal: Proc.Natl.Acad.Sci.USA / Year: 1993Title: The conformation of the sarcin/ricin loop from 28 S ribosomal RNA Authors: Szewczak, A.A. / Moore, P.B. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  480d.cif.gz 480d.cif.gz | 26.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb480d.ent.gz pdb480d.ent.gz | 17.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  480d.json.gz 480d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/80/480d https://data.pdbj.org/pub/pdb/validation_reports/80/480d ftp://data.pdbj.org/pub/pdb/validation_reports/80/480d ftp://data.pdbj.org/pub/pdb/validation_reports/80/480d | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  483dC  430dS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 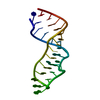
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||
| Unit cell |
|
- Components
Components
| #1: RNA chain | Mass: 8744.255 Da / Num. of mol.: 1 / Fragment: SARCIN/RICIN DOMAIN / Source method: obtained synthetically |
|---|---|
| #2: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.04 Å3/Da / Density % sol: 50 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 292 K / Method: vapor diffusion, hanging drop / pH: 7 Details: 3.0 - 3.2 M (NH4)2SO4, BUFFER X (50 MM POTASIUM MOPS, PH 7.0; 10 MM MGCL2; AND 10 MM MNCL2), AND ~2.5 MG/ML RNA, VAPOR DIFFUSION, HANGING DROP, temperature 292K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 19 ℃ / pH: 8 / Method: vapor diffusion | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 295 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 |
| Detector | Type: RIGAKU RAXIS IIC / Detector: IMAGE PLATE / Date: May 12, 1998 |
| Radiation | Monochromator: MSC MIRROR BOX / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.5→20 Å / Num. all: 10744 / % possible obs: 99.7 % / Observed criterion σ(I): -3 / Redundancy: 5.6 % / Biso Wilson estimate: 13.8 Å2 / Rmerge(I) obs: 0.052 / Net I/σ(I): 35.5 |
| Reflection shell | Resolution: 1.5→1.53 Å / Redundancy: 2.5 % / Rmerge(I) obs: 0.459 / Mean I/σ(I) obs: 2.5 / Rsym value: 0.459 / % possible all: 98.5 |
| Reflection shell | *PLUS % possible obs: 98.5 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: THE RELATED RAT SARCIN/RICIN STRUCTURE (430D) Resolution: 1.5→20 Å / Cross valid method: R FREE / σ(F): 0 / σ(I): 0 Stereochemistry target values: G. PARKINSON, J. VOJTECHOVSKY, L. CLOWNEY, A.T. BRUNGER, H.M. BERMAN: ACTA CRYST.D, 52, 57 (1996) Details: AFTER BULK SOLVENT CORRECTION AND ANISOTROPIC SCALING OF FOBS, CYCLES OF POWELL MINIMIZATION FOLLOWED BY B-FACTOR REFINEMENT WERE DONE.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 17.9 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.5→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.5→1.53 Å / Total num. of bins used: 20
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.851 / Classification: refinement X-PLOR / Version: 3.851 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 1.5 Å / Lowest resolution: 20 Å / σ(F): 0 / % reflection Rfree: 9.9 % / Rfactor obs: 0.18 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS Biso mean: 17.9 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor Rfree: 0.33 / % reflection Rfree: 7.5 % / Rfactor Rwork: 0.336 / Rfactor obs: 0.336 |
 Movie
Movie Controller
Controller





 PDBj
PDBj






























