| Entry | Database: PDB / ID: 3wx1
|
|---|
| Title | Mouse Cereblon thalidomide binding domain, selenomethionine derivative |
|---|
 Components Components | Protein cereblon |
|---|
 Keywords Keywords | METAL BINDING PROTEIN / Zinc Finger / E3 ubiquitin ligase |
|---|
| Function / homology |  Function and homology information Function and homology information
negative regulation of large conductance calcium-activated potassium channel activity / Cul4A-RING E3 ubiquitin ligase complex / locomotory exploration behavior / positive regulation of Wnt signaling pathway / negative regulation of protein-containing complex assembly / positive regulation of protein-containing complex assembly / proteasome-mediated ubiquitin-dependent protein catabolic process / transmembrane transporter binding / protein ubiquitination / perinuclear region of cytoplasm ...negative regulation of large conductance calcium-activated potassium channel activity / Cul4A-RING E3 ubiquitin ligase complex / locomotory exploration behavior / positive regulation of Wnt signaling pathway / negative regulation of protein-containing complex assembly / positive regulation of protein-containing complex assembly / proteasome-mediated ubiquitin-dependent protein catabolic process / transmembrane transporter binding / protein ubiquitination / perinuclear region of cytoplasm / metal ion binding / nucleus / membrane / cytoplasmSimilarity search - Function Peptide methionine sulfoxide reductase. / Metal Binding Protein, Guanine Nucleotide Exchange Factor; Chain A / Yippee/Mis18/Cereblon / Yippee zinc-binding/DNA-binding /Mis18, centromere assembly / CULT domain / CULT domain profile. / Lon N-terminal domain profile. / Lon protease, N-terminal domain / Lon protease, N-terminal domain superfamily / ATP-dependent protease La (LON) substrate-binding domain ...Peptide methionine sulfoxide reductase. / Metal Binding Protein, Guanine Nucleotide Exchange Factor; Chain A / Yippee/Mis18/Cereblon / Yippee zinc-binding/DNA-binding /Mis18, centromere assembly / CULT domain / CULT domain profile. / Lon N-terminal domain profile. / Lon protease, N-terminal domain / Lon protease, N-terminal domain superfamily / ATP-dependent protease La (LON) substrate-binding domain / Found in ATP-dependent protease La (LON) / PUA-like superfamily / Beta Complex / Mainly BetaSimilarity search - Domain/homology |
|---|
| Biological species |   Mus musculus (house mouse) Mus musculus (house mouse) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MAD / Resolution: 1.93 Å MAD / Resolution: 1.93 Å |
|---|
 Authors Authors | Mori, T. / Ito, T. / Hirano, Y. / Yamaguchi, Y. / Handa, H. / Hakoshima, T. |
|---|
 Citation Citation |  Journal: Nat.Struct.Mol.Biol. / Year: 2014 Journal: Nat.Struct.Mol.Biol. / Year: 2014
Title: Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs
Authors: Chamberlain, P.P. / Lopez-Girona, A. / Miller, K. / Carmel, G. / Pagarigan, B. / Chie-Leon, B. / Rychak, E. / Corral, L.G. / Ren, Y.J. / Wang, M. / Riley, M. / Delker, S.L. / Ito, T. / Ando, ...Authors: Chamberlain, P.P. / Lopez-Girona, A. / Miller, K. / Carmel, G. / Pagarigan, B. / Chie-Leon, B. / Rychak, E. / Corral, L.G. / Ren, Y.J. / Wang, M. / Riley, M. / Delker, S.L. / Ito, T. / Ando, H. / Mori, T. / Hirano, Y. / Handa, H. / Hakoshima, T. / Daniel, T.O. / Cathers, B.E. |
|---|
| History | | Deposition | Jul 10, 2014 | Deposition site: PDBJ / Processing site: PDBJ |
|---|
| Revision 1.0 | Aug 6, 2014 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Aug 27, 2014 | Group: Database references |
|---|
| Revision 1.2 | Sep 17, 2014 | Group: Database references |
|---|
| Revision 1.3 | Oct 30, 2024 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Structure summary
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_entry_details / pdbx_modification_feature / pdbx_struct_conn_angle / pdbx_struct_special_symmetry / struct_conn / struct_ref_seq_dif / struct_site
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_asym_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr2_auth_asym_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_leaving_atom_flag / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_label_asym_id / _struct_ref_seq_dif.details / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MAD / Resolution: 1.93 Å
MAD / Resolution: 1.93 Å  Authors
Authors Citation
Citation Journal: Nat.Struct.Mol.Biol. / Year: 2014
Journal: Nat.Struct.Mol.Biol. / Year: 2014 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 3wx1.cif.gz
3wx1.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb3wx1.ent.gz
pdb3wx1.ent.gz PDB format
PDB format 3wx1.json.gz
3wx1.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/wx/3wx1
https://data.pdbj.org/pub/pdb/validation_reports/wx/3wx1 ftp://data.pdbj.org/pub/pdb/validation_reports/wx/3wx1
ftp://data.pdbj.org/pub/pdb/validation_reports/wx/3wx1 Links
Links Assembly
Assembly
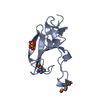

 Components
Components

 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SPring-8
SPring-8  / Beamline: BL41XU / Wavelength: 0.97911, 0.99513, 0.97941
/ Beamline: BL41XU / Wavelength: 0.97911, 0.99513, 0.97941 Processing
Processing MAD / Resolution: 1.93→34.1 Å / Cor.coef. Fo:Fc: 0.961 / Cor.coef. Fo:Fc free: 0.955 / SU B: 3.183 / SU ML: 0.092 / ESU R: 0.139 / ESU R Free: 0.125 / Stereochemistry target values: MAXIMUM LIKELIHOOD
MAD / Resolution: 1.93→34.1 Å / Cor.coef. Fo:Fc: 0.961 / Cor.coef. Fo:Fc free: 0.955 / SU B: 3.183 / SU ML: 0.092 / ESU R: 0.139 / ESU R Free: 0.125 / Stereochemistry target values: MAXIMUM LIKELIHOOD Movie
Movie Controller
Controller


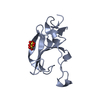

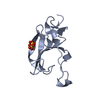








 PDBj
PDBj





