| Entry | Database: PDB / ID: 3qgl
|
|---|
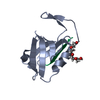
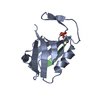
| Title | Crystal Structure of PDZ domain of sorting nexin 27 (SNX27) in complex with the ESESKV peptide corresponding to the C-terminal tail of GIRK3 |
|---|
 Components Components | - G protein-activated inward rectifier potassium channel 3
- Sorting nexin-27
|
|---|
 Keywords Keywords | PROTEIN BINDING / PDZ domain / PDZ binding / GIRK3 regulation / early endosomes / brain / neurons |
|---|
| Function / homology |  Function and homology information Function and homology information
postsynaptic early endosome / G-protein activated inward rectifier potassium channel activity / establishment of protein localization to plasma membrane / positive regulation of AMPA glutamate receptor clustering / regulation of presynaptic membrane potential / neurotransmitter receptor transport to plasma membrane / postsynaptic recycling endosome / inward rectifier potassium channel complex / WASH complex / retromer complex ...postsynaptic early endosome / G-protein activated inward rectifier potassium channel activity / establishment of protein localization to plasma membrane / positive regulation of AMPA glutamate receptor clustering / regulation of presynaptic membrane potential / neurotransmitter receptor transport to plasma membrane / postsynaptic recycling endosome / inward rectifier potassium channel complex / WASH complex / retromer complex / response to methamphetamine hydrochloride / endosome to plasma membrane protein transport / regulation of monoatomic ion transmembrane transport / phosphatidylinositol-3-phosphate binding / inward rectifier potassium channel activity / regulation of synapse maturation / endocytic recycling / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / endosomal transport / endosome to lysosome transport / potassium ion import across plasma membrane / parallel fiber to Purkinje cell synapse / immunological synapse / regulation of postsynaptic membrane neurotransmitter receptor levels / phosphatidylinositol binding / ionotropic glutamate receptor binding / PDZ domain binding / intracellular protein transport / cellular response to nerve growth factor stimulus / Schaffer collateral - CA1 synapse / calcium-dependent protein binding / nervous system development / presynaptic membrane / early endosome membrane / early endosome / endosome / postsynapse / glutamatergic synapse / signal transduction / plasma membrane / cytosolSimilarity search - Function Potassium channel, inwardly rectifying, Kir3.3 / SNX27, atypical FERM-like domain / SNX27, PX domain / SNX27, RA domain / Ras association (RalGDS/AF-6) domain / Ras-associating (RA) domain profile. / Ras-associating (RA) domain / PhoX homologous domain, present in p47phox and p40phox. / Potassium channel, inwardly rectifying, transmembrane domain / Inward rectifier potassium channel transmembrane domain ...Potassium channel, inwardly rectifying, Kir3.3 / SNX27, atypical FERM-like domain / SNX27, PX domain / SNX27, RA domain / Ras association (RalGDS/AF-6) domain / Ras-associating (RA) domain profile. / Ras-associating (RA) domain / PhoX homologous domain, present in p47phox and p40phox. / Potassium channel, inwardly rectifying, transmembrane domain / Inward rectifier potassium channel transmembrane domain / PX domain profile. / PX domain / Phox homology / PX domain superfamily / Potassium channel, inwardly rectifying, Kir, cytoplasmic / Potassium channel, inwardly rectifying, Kir / Inward rectifier potassium channel, C-terminal / Inward rectifier potassium channel C-terminal domain / PDZ domain / Pdz3 Domain / PDZ domain / PDZ domain profile. / Domain present in PSD-95, Dlg, and ZO-1/2. / PDZ domain / PDZ superfamily / Immunoglobulin E-set / Roll / Ubiquitin-like domain superfamily / Mainly BetaSimilarity search - Domain/homology |
|---|
| Biological species |   Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.31 Å MOLECULAR REPLACEMENT / Resolution: 3.31 Å |
|---|
 Authors Authors | Balana, B. / Kwiatkowski, W. / Choe, S. |
|---|
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2011 Journal: Proc.Natl.Acad.Sci.USA / Year: 2011
Title: Mechanism underlying selective regulation of G protein-gated inwardly rectifying potassium channels by the psychostimulant-sensitive sorting nexin 27.
Authors: Balana, B. / Maslennikov, I. / Kwiatkowski, W. / Stern, K.M. / Bahima, L. / Choe, S. / Slesinger, P.A. |
|---|
| History | | Deposition | Jan 24, 2011 | Deposition site: RCSB / Processing site: RCSB |
|---|
| Revision 1.0 | Mar 16, 2011 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Jul 13, 2011 | Group: Version format compliance |
|---|
| Revision 1.2 | Sep 13, 2023 | Group: Data collection / Database references / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_ref_seq_dif
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_ref_seq_dif.details |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.31 Å
MOLECULAR REPLACEMENT / Resolution: 3.31 Å  Authors
Authors Citation
Citation Journal: Proc.Natl.Acad.Sci.USA / Year: 2011
Journal: Proc.Natl.Acad.Sci.USA / Year: 2011 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 3qgl.cif.gz
3qgl.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb3qgl.ent.gz
pdb3qgl.ent.gz PDB format
PDB format 3qgl.json.gz
3qgl.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/qg/3qgl
https://data.pdbj.org/pub/pdb/validation_reports/qg/3qgl ftp://data.pdbj.org/pub/pdb/validation_reports/qg/3qgl
ftp://data.pdbj.org/pub/pdb/validation_reports/qg/3qgl

 Links
Links Assembly
Assembly





 Components
Components


 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SSRL
SSRL  / Beamline: BL9-2 / Wavelength: 0.979
/ Beamline: BL9-2 / Wavelength: 0.979  Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller










 PDBj
PDBj
